Recruitment of CRISPR-Cas systems by Tn7-like transposons
- PMID: 28811374
- PMCID: PMC5584455
- DOI: 10.1073/pnas.1709035114
Recruitment of CRISPR-Cas systems by Tn7-like transposons
Abstract
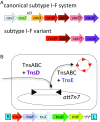
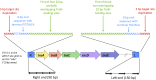
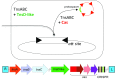
A survey of bacterial and archaeal genomes shows that many Tn7-like transposons contain minimal type I-F CRISPR-Cas systems that consist of fused cas8f and cas5f, cas7f, and cas6f genes and a short CRISPR array. Several small groups of Tn7-like transposons encompass similarly truncated type I-B CRISPR-Cas. This minimal gene complement of the transposon-associated CRISPR-Cas systems implies that they are competent for pre-CRISPR RNA (precrRNA) processing yielding mature crRNAs and target binding but not target cleavage that is required for interference. Phylogenetic analysis demonstrates that evolution of the CRISPR-Cas-containing transposons included a single, ancestral capture of a type I-F locus and two independent instances of type I-B loci capture. We show that the transposon-associated CRISPR arrays contain spacers homologous to plasmid and temperate phage sequences and, in some cases, chromosomal sequences adjacent to the transposon. We hypothesize that the transposon-encoded CRISPR-Cas systems generate displacement (R-loops) in the cognate DNA sites, targeting the transposon to these sites and thus facilitating their spread via plasmids and phages. These findings suggest the existence of RNA-guided transposition and fit the guns-for-hire concept whereby mobile genetic elements capture host defense systems and repurpose them for different stages in the life cycle of the element.
Keywords: CRISPR-Cas systems; Tn7 transposon; crRNA guide; target-site selection; transposition strategy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

