Transcriptional response to stress is pre-wired by promoter and enhancer architecture
- PMID: 28811569
- PMCID: PMC5557961
- DOI: 10.1038/s41467-017-00151-0
Transcriptional response to stress is pre-wired by promoter and enhancer architecture
Abstract
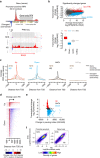
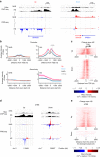
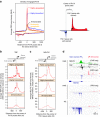
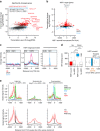
Programs of gene expression are executed by a battery of transcription factors that coordinate divergent transcription from a pair of tightly linked core initiation regions of promoters and enhancers. Here, to investigate how divergent transcription is reprogrammed upon stress, we measured nascent RNA synthesis at nucleotide-resolution, and profiled histone H4 acetylation in human cells. Our results globally show that the release of promoter-proximal paused RNA polymerase into elongation functions as a critical switch at which a gene's response to stress is determined. Highly transcribed and highly inducible genes display strong transcriptional directionality and selective assembly of general transcription factors on the core sense promoter. Heat-induced transcription at enhancers, instead, correlates with prior binding of cell-type, sequence-specific transcription factors. Activated Heat Shock Factor 1 (HSF1) binds to transcription-primed promoters and enhancers, and CTCF-occupied, non-transcribed chromatin. These results reveal chromatin architectural features that orient transcription at divergent regulatory elements and prime transcriptional responses genome-wide.Heat Shock Factor 1 (HSF1) is a regulator of stress-induced transcription. Here, the authors investigate changes to transcription and chromatin organization upon stress and find that activated HSF1 binds to transcription-primed promoters and enhancers, and to CTCF occupied, untranscribed chromatin.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

