Comparative Genome Analysis of Rathayibacter tritici NCPPB 1953 with Rathayibacter toxicus Strains Can Facilitate Studies on Mechanisms of Nematode Association and Host Infection
- PMID: 28811754
- PMCID: PMC5538441
- DOI: 10.5423/PPJ.OA.01.2017.0017
Comparative Genome Analysis of Rathayibacter tritici NCPPB 1953 with Rathayibacter toxicus Strains Can Facilitate Studies on Mechanisms of Nematode Association and Host Infection
Abstract
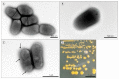
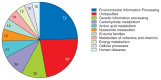
Rathayibacter tritici, which is a Gram positive, plant pathogenic, non-motile, and rod-shaped bacterium, causes spike blight in wheat and barley. For successful pathogenesis, R. tritici is associated with Anguina tritici, a nematode, which produces seed galls (ear cockles) in certain plant varieties and facilitates spread of infection. Despite significant efforts, little research is available on the mechanism of disease or bacteria-nematode association of this bacterium due to lack of genomic information. Here, we report the first complete genome sequence of R. tritici NCPPB 1953 with diverse features of this strain. The whole genome consists of one circular chromosome of 3,354,681 bp with a GC content of 69.48%. A total of 2,979 genes were predicted, comprising 2,866 protein coding genes and 49 RNA genes. The comparative genomic analyses between R. tritici NCPPB 1953 and R. toxicus strains identified 1,052 specific genes in R. tritici NCPPB 1953. Using the BlastKOALA database, we revealed that the flexible genome of R. tritici NCPPB 1953 is highly enriched in 'Environmental Information Processing' system and metabolic processes for diverse substrates. Furthermore, many specific genes of R. tritici NCPPB 1953 are distributed in substrate-binding proteins for extracellular signals including saccharides, lipids, phosphates, amino acids and metallic cations. These data provides clues on rapid and stable colonization of R. tritici for disease mechanism and nematode association.
Keywords: Rathayibacter toxicus; Rathayibacter tritici NCPPB 1953; comparative genome.
Figures




References
-
- Akhtar MA. Outbreaks and new records. Pakistan. Bacterial gumming disease of wheat. FAO Plant Prot Bull. 1987;35:102.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

