A Circulating MicroRNA Signature Serves as a Diagnostic and Prognostic Indicator in Sarcoidosis
- PMID: 28812922
- PMCID: PMC5941311
- DOI: 10.1165/rcmb.2017-0207OC
A Circulating MicroRNA Signature Serves as a Diagnostic and Prognostic Indicator in Sarcoidosis
Abstract
MicroRNAs (miRNAs) act as post-transcriptional regulators of gene expression. In sarcoidosis, aberrant miRNA expression may enhance immune responses mounted against an unknown antigenic agent. We tested whether a distinct miRNA signature functions as a diagnostic biomarker and explored its role as an immune modulator in sarcoidosis. The expression of miRNAs in peripheral blood mononuclear cells from subjects who met clinical and histopathologic criteria for sarcoidosis was compared with that observed in matched controls in the ACCESS (A Case Controlled Etiologic Study of Sarcoidosis) study. Signature miRNAs were determined by miRNA microarray analysis and validated by quantitative RT-PCR. Microarray analysis identified 54 mature, human feature miRNAs that were differentially expressed between the groups. Significant feature miRNAs that distinguished subjects with sarcoidosis from controls were selected by means of probabilistic models adjusted for clinical variables. Eight signature miRNAs were chosen to verify the diagnosis of sarcoidosis in a validation cohort, and distinguished subjects with sarcoidosis from controls with a positive predictive value of 88%. We identified both novel and previously described genes and molecular pathways associated with sarcoidosis as targets of these signature miRNAs. Additionally, we demonstrate that signature miRNAs (hsa-miR-150-3p and hsa-miR-342-5p) are significantly associated with reduced lymphocytes and airflow limitations, both of which are known markers of a poor prognosis. Together, these findings suggest that a circulating miRNA signature serves as a noninvasive biomarker that supports the diagnosis of sarcoidosis. Future studies will test the miRNA signature as a prognostication tool to identify unfavorable changes associated with poor clinical outcomes in sarcoidosis.
Keywords: biomarker; gene regulatory network; miRNA; peripheral blood mononuclear cells; sarcoidosis.
Figures

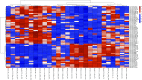

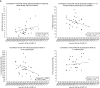

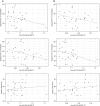


References
-
- Grunewald J, Spagnolo P, Wahlström J, Eklund A. Immunogenetics of disease-causing inflammation in sarcoidosis. Clin Rev Allergy Immunol. 2015;49:19–35. - PubMed
-
- Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. - PubMed
-
- Hunninghake GW, Costabel U, Ando M, Baughman R, Cordier JF, du Bois R, et al. ATS/ERS/WASOG statement on sarcoidosis. American Thoracic Society/European Respiratory Society/World Association of Sarcoidosis and other Granulomatous Disorders. Sarcoidosis Vasc Diffuse Lung Dis. 1999;16:149–173. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

