Celebrating wobble decoding: Half a century and still much is new
- PMID: 28812932
- PMCID: PMC6103715
- DOI: 10.1080/15476286.2017.1356562
Celebrating wobble decoding: Half a century and still much is new
Abstract
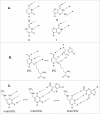
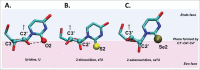
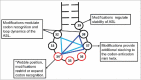
A simple post-transcriptional modification of tRNA, deamination of adenosine to inosine at the first, or wobble, position of the anticodon, inspired Francis Crick's Wobble Hypothesis 50 years ago. Many more naturally-occurring modifications have been elucidated and continue to be discovered. The post-transcriptional modifications of tRNA's anticodon domain are the most diverse and chemically complex of any RNA modifications. Their contribution with regards to chemistry, structure and dynamics reveal individual and combined effects on tRNA function in recognition of cognate and wobble codons. As forecast by the Modified Wobble Hypothesis 25 years ago, some individual modifications at tRNA's wobble position have evolved to restrict codon recognition whereas others expand the tRNA's ability to read as many as four synonymous codons. Here, we review tRNA wobble codon recognition using specific examples of simple and complex modification chemistries that alter tRNA function. Understanding natural modifications has inspired evolutionary insights and possible innovation in protein synthesis.
Keywords: Modified Wobble Hypothesis; Wobble Hypothesis; cognate and wobble codon recognition; modified nucleosides; nucleoside tautomers; tRNA; translation; wobble decoding; wobble nucleoside.
Figures

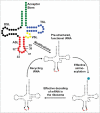
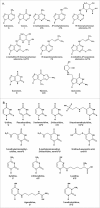

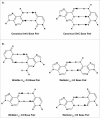
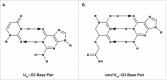


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
