Plasma metabolomics reveals membrane lipids, aspartate/asparagine and nucleotide metabolism pathway differences associated with chloroquine resistance in Plasmodium vivax malaria
- PMID: 28813452
- PMCID: PMC5559093
- DOI: 10.1371/journal.pone.0182819
Plasma metabolomics reveals membrane lipids, aspartate/asparagine and nucleotide metabolism pathway differences associated with chloroquine resistance in Plasmodium vivax malaria
Abstract
Background: Chloroquine (CQ) is the main anti-schizontocidal drug used in the treatment of uncomplicated malaria caused by Plasmodium vivax. Chloroquine resistant P. vivax (PvCR) malaria in the Western Pacific region, Asia and in the Americas indicates a need for biomarkers of resistance to improve therapy and enhance understanding of the mechanisms associated with PvCR. In this study, we compared plasma metabolic profiles of P. vivax malaria patients with PvCR and chloroquine sensitive parasites before treatment to identify potential molecular markers of chloroquine resistance.
Methods: An untargeted high-resolution metabolomics analysis was performed on plasma samples collected in a malaria clinic in Manaus, Brazil. Male and female patients with Plasmodium vivax were included (n = 46); samples were collected before CQ treatment and followed for 28 days to determine PvCR, defined as the recurrence of parasitemia with detectable plasma concentrations of CQ ≥100 ng/dL. Differentially expressed metabolic features between CQ-Resistant (CQ-R) and CQ-Sensitive (CQ-S) patients were identified using partial least squares discriminant analysis and linear regression after adjusting for covariates and multiple testing correction. Pathway enrichment analysis was performed using Mummichog.
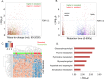
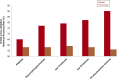
Results: Linear regression and PLS-DA methods yielded 69 discriminatory features between CQ-R and CQ-S groups, with 10-fold cross-validation classification accuracy of 89.6% using a SVM classifier. Pathway enrichment analysis showed significant enrichment (p<0.05) of glycerophospholipid metabolism, glycosphingolipid metabolism, aspartate and asparagine metabolism, purine and pyrimidine metabolism, and xenobiotics metabolism. Glycerophosphocholines levels were significantly lower in the CQ-R group as compared to CQ-S patients and also to independent control samples.
Conclusions: The results show differences in lipid, amino acids, and nucleotide metabolism pathways in the plasma of CQ-R versus CQ-S patients prior to antimalarial treatment. Metabolomics phenotyping of P. vivax samples from patients with well-defined clinical CQ-resistance is promising for the development of new tools to understand the biological process and to identify potential biomarkers of PvCR.
Conflict of interest statement
Figures


References
-
- Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis. 2012;6(9):e1814 Epub 2012/09/13. doi: 10.1371/journal.pntd.0001814 ; PubMed Central PMCID: PMC3435256. - DOI - PMC - PubMed
-
- WHO. Malária Report 2016. 2016.
-
- Tjitra E, Anstey NM, Sugiarto P, Warikar N, Kenangalem E, Karyana M, et al. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5(6):e128 Epub 2008/06/20. doi: 10.1371/journal.pmed.0050128 ; PubMed Central PMCID: PMC2429950. - DOI - PMC - PubMed
-
- Genton B, D'Acremont V, Rare L, Baea K, Reeder JC, Alpers MP, et al. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008;5(6):e127 Epub 2008/06/20. doi: 10.1371/journal.pmed.0050127 ; PubMed Central PMCID: PMC2429951. - DOI - PMC - PubMed
-
- Siqueira AM, Lacerda MV, Magalhaes BM, Mourao MP, Melo GC, Alexandre MA, et al. Characterization of Plasmodium vivax-associated admissions to reference hospitals in Brazil and India. BMC medicine. 2015;13:57 Epub 2015/04/19. doi: 10.1186/s12916-015-0302-y ; PubMed Central PMCID: PMC4404636. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

