Whole-Exome Sequencing Identifies Novel Variants for Tooth Agenesis
- PMID: 28813618
- PMCID: PMC6728545
- DOI: 10.1177/0022034517724149
Whole-Exome Sequencing Identifies Novel Variants for Tooth Agenesis
Abstract
Tooth agenesis is a common craniofacial abnormality in humans and represents failure to develop 1 or more permanent teeth. Tooth agenesis is complex, and variations in about a dozen genes have been reported as contributing to the etiology. Here, we combined whole-exome sequencing, array-based genotyping, and linkage analysis to identify putative pathogenic variants in candidate disease genes for tooth agenesis in 10 multiplex Turkish families. Novel homozygous and heterozygous variants in LRP6, DKK1, LAMA3, and COL17A1 genes, as well as known variants in WNT10A, were identified as likely pathogenic in isolated tooth agenesis. Novel variants in KREMEN1 were identified as likely pathogenic in 2 families with suspected syndromic tooth agenesis. Variants in more than 1 gene were identified segregating with tooth agenesis in 2 families, suggesting oligogenic inheritance. Structural modeling of missense variants suggests deleterious effects to the encoded proteins. Functional analysis of an indel variant (c.3607+3_6del) in LRP6 suggested that the predicted resulting mRNA is subject to nonsense-mediated decay. Our results support a major role for WNT pathways genes in the etiology of tooth agenesis while revealing new candidate genes. Moreover, oligogenic cosegregation was suggestive for complex inheritance and potentially complex gene product interactions during development, contributing to improved understanding of the genetic etiology of familial tooth agenesis.
Keywords: WNT signaling pathway; array genotyping; gene; hypodontia; next generation sequencing; oligodontia.
Conflict of interest statement
J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron, and is a co-inventor on multiple U.S. and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The other authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures


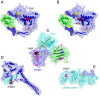
References
-
- The ARIC investigators. 1989. The atherosclerosis risk in communities (ARIC) study: design and objectives. Am J Epidemiol. 129(4):687–702. - PubMed
-
- Chen Y, Bei M, Woo I, Satokata I, Maas R. 1996. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 122(10):3035–3044. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

