The DNA Methylcytosine Dioxygenase Tet2 Sustains Immunosuppressive Function of Tumor-Infiltrating Myeloid Cells to Promote Melanoma Progression
- PMID: 28813659
- PMCID: PMC5710009
- DOI: 10.1016/j.immuni.2017.07.020
The DNA Methylcytosine Dioxygenase Tet2 Sustains Immunosuppressive Function of Tumor-Infiltrating Myeloid Cells to Promote Melanoma Progression
Abstract
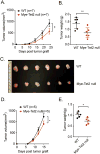
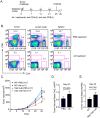
Ten-Eleven-Translocation-2 (Tet2) is a DNA methylcytosine dioxygenase that functions as a tumor suppressor in hematopoietic malignancies. We examined the role of Tet2 in tumor-tissue myeloid cells and found that Tet2 sustains the immunosuppressive function of these cells. We found that Tet2 expression is increased in intratumoral myeloid cells both in mouse models of melanoma and in melanoma patients and that this increased expression is dependent on an IL-1R-MyD88 pathway. Ablation of Tet2 in myeloid cells suppressed melanoma growth in vivo and shifted the immunosuppressive gene expression program in tumor-associated macrophages to a proinflammatory one, with a concomitant reduction of the immunosuppressive function. This resulted in increased numbers of effector T cells in the tumor, and T cell depletion abolished the reduced tumor growth observed upon myeloid-specific deletion of Tet2. Our findings reveal a non-cell-intrinsic, tumor-promoting function for Tet2 and suggest that Tet2 may present a therapeutic target for the treatment of non-hematologic malignancies.
Keywords: 5hmC; 5mC; Arg1; BMDM; HSC; Il1b; MDSC; TAM; Tet3; YUMM1.7.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nature immunology. 2016;17:18–25. - PubMed
-
- Bhatia S, Thompson JA. Melanoma: immune checkpoint blockade story gets better. Lancet. 2014;384:1078–1079. - PubMed
-
- Biswas SK, Sica A, Lewis CE. Plasticity of macrophage function during tumor progression: regulation by distinct molecular mechanisms. J Immunol. 2008;180:2011–2017. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

