Zygotic Genome Activation Occurs Shortly after Fertilization in Maize
- PMID: 28814645
- PMCID: PMC5635985
- DOI: 10.1105/tpc.17.00099
Zygotic Genome Activation Occurs Shortly after Fertilization in Maize
Abstract
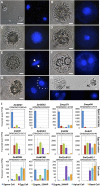
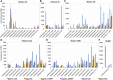
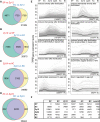
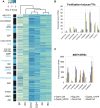
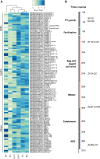
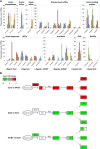
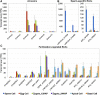
The formation of a zygote via the fusion of an egg and sperm cell and its subsequent asymmetric division herald the start of the plant's life cycle. Zygotic genome activation (ZGA) is thought to occur gradually, with the initial steps of zygote and embryo development being primarily maternally controlled, and subsequent steps being governed by the zygotic genome. Here, using maize (Zea mays) as a model plant system, we determined the timing of zygote development and generated RNA-seq transcriptome profiles of gametes, zygotes, and apical and basal daughter cells. ZGA occurs shortly after fertilization and involves ∼10% of the genome being activated in a highly dynamic pattern. In particular, genes encoding transcriptional regulators of various families are activated shortly after fertilization. Further analyses suggested that chromatin assembly is strongly modified after fertilization, that the egg cell is primed to activate the translational machinery, and that hormones likely play a minor role in the initial steps of early embryo development in maize. Our findings provide important insights into gamete and zygote activity in plants, and our RNA-seq transcriptome profiles represent a comprehensive, unique RNA-seq data set that can be used by the research community.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures

References
-
- Ahmad K., Henikoff S. (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191–1200. - PubMed
-
- Anderson S.N., Johnson C.S., Jones D.S., Conrad L.J., Gou X., Russell S.D., Sundaresan V. (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 76: 729–741. - PubMed
-
- Autran D., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719. - PubMed
-
- Baroux C., Grossniklaus U. (2015). The maternal-to-zygotic transition in flowering plants: Evidence, mechanisms, and plasticity. Curr. Top. Dev. Biol. 113: 351–371. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

