Adaptive simulations, towards interactive protein-ligand modeling
- PMID: 28814780
- PMCID: PMC5559483
- DOI: 10.1038/s41598-017-08445-5
Adaptive simulations, towards interactive protein-ligand modeling
Abstract
Modeling the dynamic nature of protein-ligand binding with atomistic simulations is one of the main challenges in computational biophysics, with important implications in the drug design process. Although in the past few years hardware and software advances have significantly revamped the use of molecular simulations, we still lack a fast and accurate ab initio description of the binding mechanism in complex systems, available only for up-to-date techniques and requiring several hours or days of heavy computation. Such delay is one of the main limiting factors for a larger penetration of protein dynamics modeling in the pharmaceutical industry. Here we present a game-changing technology, opening up the way for fast reliable simulations of protein dynamics by combining an adaptive reinforcement learning procedure with Monte Carlo sampling in the frame of modern multi-core computational resources. We show remarkable performance in mapping the protein-ligand energy landscape, being able to reproduce the full binding mechanism in less than half an hour, or the active site induced fit in less than 5 minutes. We exemplify our method by studying diverse complex targets, including nuclear hormone receptors and GPCRs, demonstrating the potential of using the new adaptive technique in screening and lead optimization studies.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

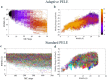


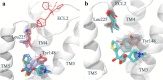

References
-
- Orozco, M. A theoretical view of protein dynamics. Chem Soc Rev (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

