Evolutionary history of the mariner element galluhop in avian genomes
- PMID: 28814978
- PMCID: PMC5556988
- DOI: 10.1186/s13100-017-0094-z
Evolutionary history of the mariner element galluhop in avian genomes
Abstract
Background: Transposable elements (TEs) are highly abundant genomic parasites in eukaryote genomes. Although several genomes have been screened for TEs, so far very limited information is available regarding avian TEs and their evolutionary histories. Taking advantage of the rich genomic data available for birds, we characterized the evolutionary history of the galluhop element, originally described in Gallus gallus, through the use of several bioinformatic analyses.
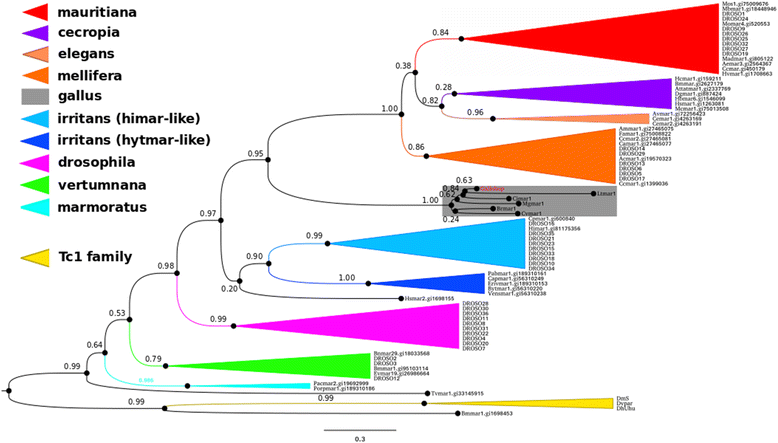
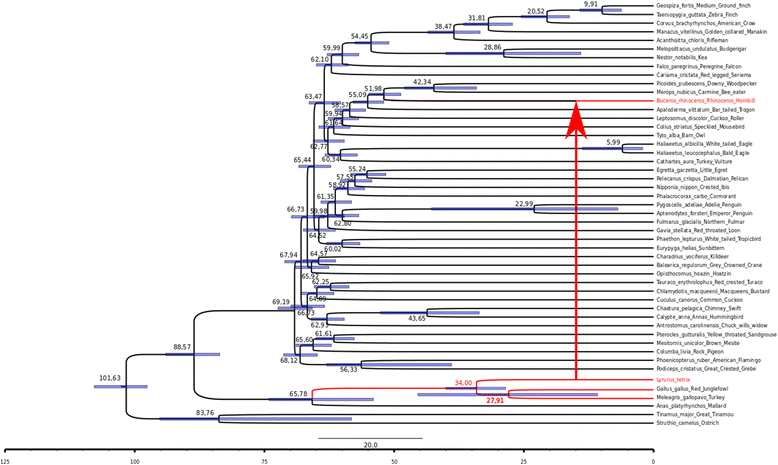
Results: galluhop homologous sequences were found in 6 of 72 genomes analyzed: 5 species of Galliformes (Gallus gallus, Meleagris gallopavo, Coturnix japonica, Colinus virginianus, Lyrurus tetrix) and one Buceritiformes (Buceros rhinoceros). The copy number ranged from 5 to 10,158, in the genomes of C. japonica and G. gallus respectively. All 6 species possessed short elements, suggesting the presence of Miniature Inverted repeats Transposable Elements (MITEs), which underwent an ancient massive amplification in the G. gallus and M. gallopavo genomes. Only 4 species showed potential MITE full-length partners, although no potential coding copies were detected. Phylogenetic analysis of reconstructed coding sequences showed that galluhop homolog sequences form a new mariner subfamily, which we termed Gallus. Inter-species and intragenomic galluhop distance analyses indicated a high identity between the consensus of B. rhinoceros and the other 5 related species, and different emergence ages of the element between the Galliformes species and B. rhinocerus, suggesting that horizontal transfer took place from Galliformes to a Buceritiformes ancestor, probably through an intermediate species.
Conclusions: Overall, our results showed that mariner elements have amplified to high copy numbers in some avian species, and that this transposition burst probably occurred in the common ancestor of G. gallus and M. gallopavo. In addition, although no coding sequences could be found currently, they probably existed, allowing an ancient massive MITE amplification in these 2 species. The other 4 species also have MITEs, suggesting that this new mariner family is prone to give rise to such non-autonomous derivatives. Last, our results suggest that a horizontal transfer event of a galluhop element occurred between Galliformes and Buceritiformes.
Keywords: Avian genome; Galluhop; Genomic parasites; Horizontal transfer; MITEs; Mariner.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989. [cited 1989 Apr 1];5:103–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/2543105. - PubMed
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. [Internet]. Nature Publishing Group; 2006 [cited 2006];8:973–82. Available from: http://www.nature.com/nrg/journal/vaop/ncurrent/full/nrg2165.html. - PubMed
-
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008 [cited 2008 May 1];9:411–2; author reply 414. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18421312. - PubMed
-
- Kidwell MGMG, Lisch DRDR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evol Int J Org Evol. 2000 [cited 2000];55:1–24. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11263730. - PubMed
-
- Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004 [cited 2004 Jan 1];6:57–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14632259. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

