The Role of Epigenetics in Type 1 Diabetes
- PMID: 28815391
- PMCID: PMC5559569
- DOI: 10.1007/s11892-017-0916-x
The Role of Epigenetics in Type 1 Diabetes
Abstract
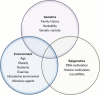
Purpose of review: Epigenetics is defined as mitotically heritable changes in gene expression that do not directly alter the DNA sequence. By implication, such epigenetic changes are non-genetically determined, although they can be affected by inherited genetic variation. Extensive evidence indicates that autoimmune diseases including type 1 diabetes are determined by the interaction of genetic and non-genetic factors. Much is known of the genetic causes of these diseases, but the non-genetic effects are less clear-cut. Further, it remains unclear how they interact to cause the destructive autoimmune process. This review identifies the key issues in the genetic/non-genetic interaction, examining the most recent evidence of the role of non-genetic effects in the disease process, including the impact of epigenetic effects on key pathways.
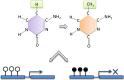
Recent findings: Recent research indicates that these pathways likely involve immune effector cells both of the innate and adaptive immune response. Specifically, there is evidence of cell type-specific enrichment in altered DNA methylation, changes which were temporally stable and enriched at gene regulatory elements. Epigenomics remains in its infancy, and we anticipate further studies will define how the interaction of genetic and non-genetic effects induces tissue-specific destruction and enhances our ability to predict, and possibly even modify that process.
Keywords: Diabetes; Epigenetics; Methylation; Type 1.
Conflict of interest statement
Conflict of Interest
Samuel T. Jerram, Mary N. Dang and R. David Leslie declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

