Development of a silicon limitation inducible expression system for recombinant protein production in the centric diatoms Thalassiosira pseudonana and Cyclotella cryptica
- PMID: 28818078
- PMCID: PMC5561644
- DOI: 10.1186/s12934-017-0760-3
Development of a silicon limitation inducible expression system for recombinant protein production in the centric diatoms Thalassiosira pseudonana and Cyclotella cryptica
Abstract
Background: An inducible promoter for recombinant protein expression provides substantial benefits because under induction conditions cellular energy and metabolic capability can be directed into protein synthesis. The most widely used inducible promoter for diatoms is for nitrate reductase, however, nitrogen metabolism is tied into diverse aspects of cellular function, and the induction response is not necessarily robust. Silicon limitation offers a means to eliminate energy and metabolic flux into cell division processes, with little other detrimental effect on cellular function, and a protein expression system that works under those conditions could be advantageous.
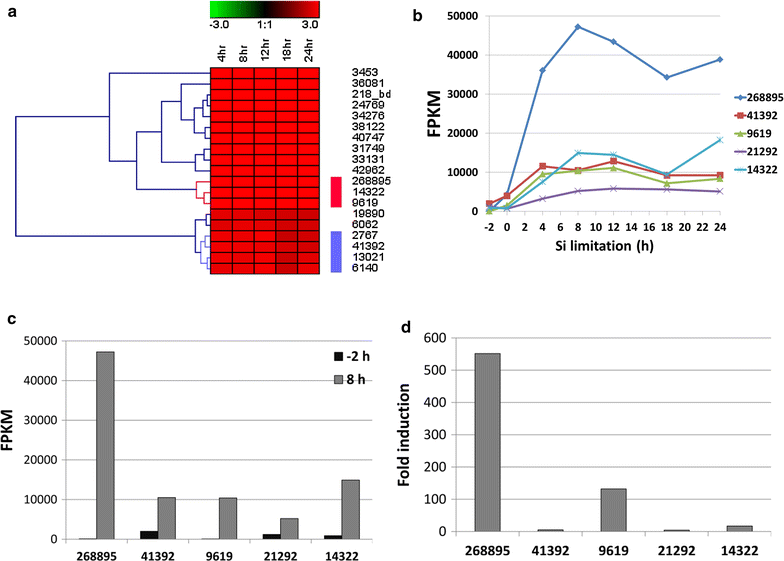
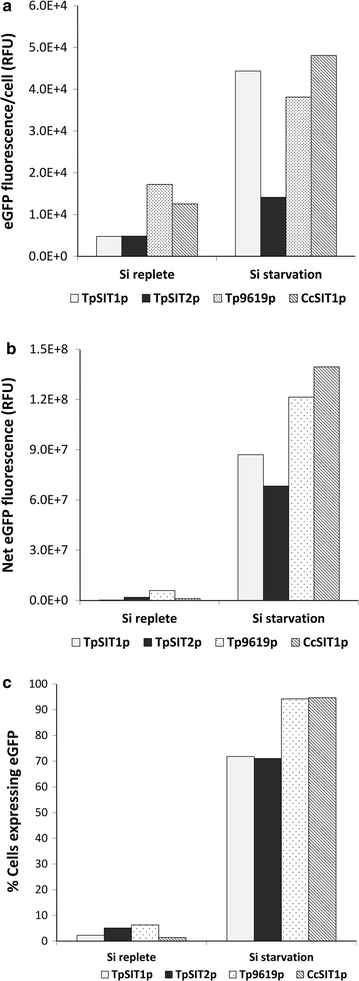
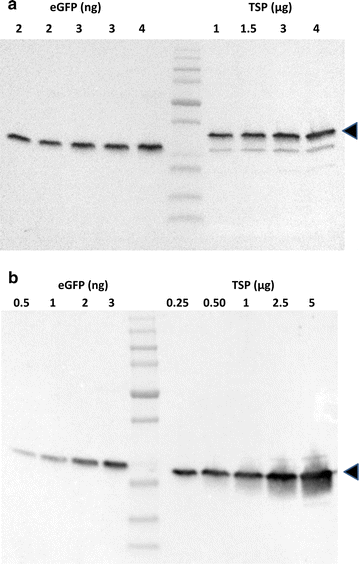
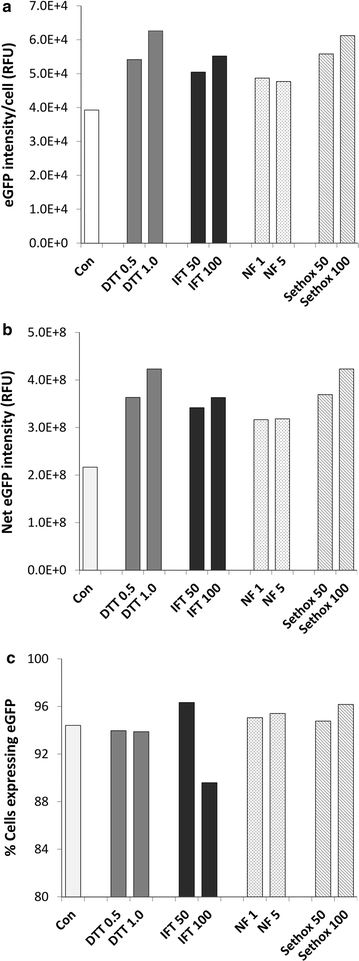
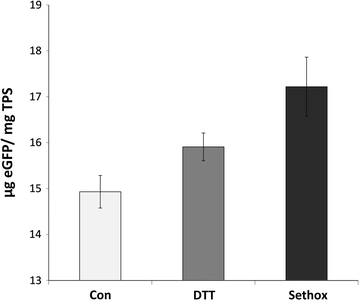
Results: In this study, we evaluate a number of promoters for recombinant protein expression induced by silicon limitation and repressed by the presence of silicon in the diatoms Thalassiosira pseudonana and Cyclotella cryptica. In addition to silicon limitation, we describe additional strategies to elevate recombinant protein expression level, including inclusion of the 5' fragment of the coding region of the native gene and reducing carbon flow into ancillary processes of pigment synthesis and formation of photosynthetic storage products. We achieved yields of eGFP to 1.8% of total soluble protein in C. cryptica, which is about 3.6-fold higher than that obtained with chloroplast expression and ninefold higher than nuclear expression in another well-established algal system.
Conclusions: Our studies demonstrate that the combination of inducible promoter and other strategies can result in robust expression of recombinant protein in a nuclear-based expression system in diatoms under silicon limited conditions, separating the protein expression regime from growth processes and improving overall recombinant protein yields.
Keywords: Cyclotella cryptica; Diatom; Fatty acid synthesis inhibition; Inducible promoter; Protein expression; Silicon transporter; Silicon-limitation; Thalassiosira pseudonana; Transformation.
Figures




References
-
- Sevastsyanovich YR, Leyton DL, Wells TJ, Wardius CA, Tveen-Jensen K, Morris FC, Knowles TJ, Cunningham AF, Cole JA, Henderson IR. A generalised module for the selective extracellular accumulation of recombinant proteins. Microb Cell Fact. 2012;11:69. doi: 10.1186/1475-2859-11-69. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

