CD24 induces changes to the surface receptors of B cell microvesicles with variable effects on their RNA and protein cargo
- PMID: 28819186
- PMCID: PMC5561059
- DOI: 10.1038/s41598-017-08094-8
CD24 induces changes to the surface receptors of B cell microvesicles with variable effects on their RNA and protein cargo
Abstract
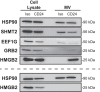
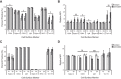
The CD24 cell surface receptor promotes apoptosis in developing B cells, and we recently found that it induces B cells to release plasma membrane-derived, CD24-bearing microvesicles (MVs). Here we have performed a systematic characterization of B cell MVs released from WEHI-231 B lymphoma cells in response to CD24 stimulation. We found that B cells constitutively release MVs of approximately 120 nm, and that CD24 induces an increase in phosphatidylserine-positive MV release. RNA cargo is predominantly comprised of 5S rRNA, regardless of stimulation; however, CD24 causes a decrease in the incorporation of protein coding transcripts. The MV proteome is enriched with mitochondrial and metabolism-related proteins after CD24 stimulation; however, these changes were variable and could not be fully validated by Western blotting. CD24-bearing MVs carry Siglec-2, CD63, IgM, and, unexpectedly, Ter119, but not Siglec-G or MHC-II despite their presence on the cell surface. CD24 stimulation also induces changes in CD63 and IgM expression on MVs that is not mirrored by the changes in cell surface expression. Overall, the composition of these MVs suggests that they may be involved in releasing mitochondrial components in response to pro-apoptotic stress with changes to the surface receptors potentially altering the cell type(s) that interact with the MVs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Müller G. Release of exosomes and microvesicles harbouring specific RNAs and glycosylphosphatidylinositol-anchored proteins from rat and human adipocytes is controlled by histone methylation. American Journal of Molecular Biology. 2012;02:187–209. doi: 10.4236/ajmb.2012.23020. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

