Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin
- PMID: 28819201
- PMCID: PMC5561251
- DOI: 10.1038/s41467-017-00338-5
Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin
Abstract
Linker histone H1 is an important structural component of chromatin that stabilizes the nucleosome and compacts the nucleofilament into higher-order structures. The biology of histone H1 remains, however, poorly understood. Here we show that Drosophila histone H1 (dH1) prevents genome instability as indicated by the increased γH2Av (H2AvS137P) content and the high incidence of DNA breaks and sister-chromatid exchanges observed in dH1-depleted cells. Increased γH2Av occurs preferentially at heterochromatic elements, which are upregulated upon dH1 depletion, and is due to the abnormal accumulation of DNA:RNA hybrids (R-loops). R-loops accumulation is readily detectable in G1-phase, whereas γH2Av increases mainly during DNA replication. These defects induce JNK-mediated apoptosis and are specific of dH1 depletion since they are not observed when heterochromatin silencing is relieved by HP1a depletion. Altogether, our results suggest that histone H1 prevents R-loops-induced DNA damage in heterochromatin and unveil its essential contribution to maintenance of genome stability.While structural importance of linker histone H1 in packaging eukaryotic genome into chromatin is well known, its biological function remains poorly understood. Here the authors reveal that Drosophila linker histone H1 prevents DNA:RNA hybrids accumulation and genome instability in heterochromatin.
Conflict of interest statement
The authors declare no competing financial interests.
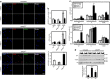
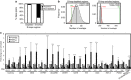
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

