The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols
- PMID: 28819242
- PMCID: PMC5561025
- DOI: 10.1038/s41598-017-07885-3
The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols
Abstract
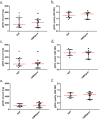
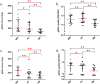
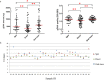
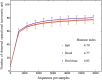
Saliva has attracted attention as a diagnostic fluid due to the association of oral microbiota with systemic diseases. However, the lack of standardised methods for saliva collection has led to the slow uptake of saliva in microbiome research. The aim of this study was to systematically evaluate the potential effects on salivary microbiome profiles using different methods of saliva collection, storage and gDNA extraction. Three types of saliva fractions were collected from healthy individuals with or without the gDNA stabilising buffer. Subsequently, three types of gDNA extraction methods were evaluated to determine the gDNA extraction efficiencies from saliva samples. The purity of total bacterial gDNA was evaluated using the ratio of human β-globin to bacterial 16S rRNA PCR while 16S rRNA gene amplicon sequencing was carried out to identify the bacterial profiles present in these samples. The quantity and quality of extracted gDNA were similar among all three gDNA extraction methods and there were no statistically significant differences in the bacterial profiles among different saliva fractions at the genus-level of taxonomic classification. In conclusion, saliva sampling, processing and gDNA preparation do not have major influence on microbiome profiles.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- na Head and Neck Cancer Biomarkers Detected in Saliva. Cancer Biol Ther4, 6–12 (2005).
-
- Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J. Proteome Res. 2009;8:5590–5600. doi: 10.1021/pr900675w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

