Sources to variability in circulating human miRNA signatures
- PMID: 28820329
- PMCID: PMC5731815
- DOI: 10.1080/15476286.2017.1367888
Sources to variability in circulating human miRNA signatures
Abstract
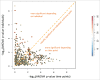
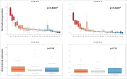
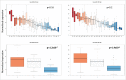
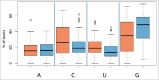
An increasing number of studies propose circulating microRNAs (miRNAs) as biomarkers for a large number of human diseases including cancer, cardiovascular diseases, neurologic pathologies and others. To further validate miRNA as biomarkers it is indispensable to understand the variability of circulating miRNAs in healthy individuals. We determined the longitudinal miRNomes of 90 serum samples from the Janus Serum Bank in Norway, which have been stored between 23 and 40 y at -25 °Celsius. We profiled 3 serum samples with microarrays for 30 individuals, each. For each individual the samples were collected with a time interval of approximately 5 y. This design allowed insights into inter-individual variability, age dependent miRNA variability and the impact of storage length and pre-processing. A significant proportion of the miRNome was affected by the age of the blood donor and a not negligible, albeit small, part of the miRNome by the storage time. A substantial part of miRNAs was differentially abundant between individuals, independent of the time when samples were collected. Stepwise filtering of the 529 miRNAs that were detected in the serum samples showed 168 miRNAs with differential abundance depending on the time point analyzed, 56 miRNAs differentially abundant between individuals, and 169 miRNAs with an abundance depending on the sampling procedure. While these groups of miRNAs contain generally interesting and biologically important miRNAs, the remaining 135 miRNAs constitute very promising biomarker candidates as they show an overall low variability between healthy individuals, a likewise overall low variability across a longer life span, and a high independence of the sampling process and the storage length.
Keywords: circulating biomarkers; control signatures; miRNA; serum.
Figures




References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
