Accumulation of Deleterious Mutations During Bacterial Range Expansions
- PMID: 28821588
- PMCID: PMC5629331
- DOI: 10.1534/genetics.117.300144
Accumulation of Deleterious Mutations During Bacterial Range Expansions
Abstract
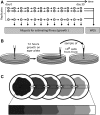
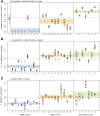
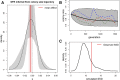
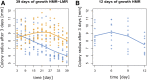
Recent theory predicts that the fitness of pioneer populations can decline when species expand their range, due to high rates of genetic drift on wave fronts making selection less efficient at purging deleterious variants. To test these predictions, we studied the fate of mutator bacteria expanding their range for 1650 generations on agar plates. In agreement with theory, we find that growth abilities of strains with a high mutation rate (HMR lines) decreased significantly over time, unlike strains with a lower mutation rate (LMR lines) that present three to four times fewer mutations. Estimation of the distribution of fitness effect under a spatially explicit model reveals a mean negative effect for new mutations (-0.38%), but it suggests that both advantageous and deleterious mutations have accumulated during the experiment. Furthermore, the fitness of HMR lines measured in different environments has decreased relative to the ancestor strain, whereas that of LMR lines remained unchanged. Contrastingly, strains with a HMR evolving in a well-mixed environment accumulated less mutations than agar-evolved strains and showed an increased fitness relative to the ancestor. Our results suggest that spatially expanding species are affected by deleterious mutations, leading to a drastic impairment of their evolutionary potential.
Keywords: experimental evolution; mutation load; range expansions.
Copyright © 2017 by the Genetics Society of America.
Figures



References
-
- Agrawal A. F., Whitlock M. C., 2012. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst. 43: 115–135.
-
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247. - PubMed
-
- Blundell J., Levy S., Venkataram S., Petrov D., Fisher D., et al. , 2015. Quantitative evolutionary dynamics of one million barcoded lineages. Bull. Am. Phys. Soc. Volume 60.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

