Dsc E3 ligase localization to the Golgi requires the ATPase Cdc48 and cofactor Ufd1 for activation of sterol regulatory element-binding protein in fission yeast
- PMID: 28821619
- PMCID: PMC5625062
- DOI: 10.1074/jbc.M117.802025
Dsc E3 ligase localization to the Golgi requires the ATPase Cdc48 and cofactor Ufd1 for activation of sterol regulatory element-binding protein in fission yeast
Abstract
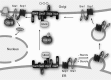
Sterol regulatory element-binding proteins (SREBPs) in the fission yeast Schizosaccharomyces pombe regulate lipid homeostasis and the hypoxic response under conditions of low sterol or oxygen availability. SREBPs are cleaved in the Golgi through the combined action of the Dsc E3 ligase complex, the rhomboid protease Rbd2, and the essential ATPases associated with diverse cellular activities (AAA+) ATPase Cdc48. The soluble SREBP N-terminal transcription factor domain is then released into the cytosol to enter the nucleus and regulate gene expression. Previously, we reported that Cdc48 binding to Rbd2 is required for Rbd2-mediated SREBP cleavage. Here, using affinity chromatography and mass spectrometry experiments, we identified Cdc48-binding proteins in S. pombe, generating a list of many previously unknown potential Cdc48-binding partners. We show that the established Cdc48 cofactor Ufd1 is required for SREBP cleavage but does not interact with the Cdc48-Rbd2 complex. Cdc48-Ufd1 is instead required at a step prior to Rbd2 function, during Golgi localization of the Dsc E3 ligase complex. Together, these findings demonstrate that two distinct Cdc48 complexes, Cdc48-Ufd1 and Cdc48-Rbd2, are required for SREBP activation and low-oxygen adaptation in S. pombe.
Keywords: ATPases associated with diverse cellular activities (AAA); Cdc48; E3 ubiquitin ligase; SREBP; Schizosaccharomyces pombe; VCP; hypoxia; membrane transport; p97; transcription regulation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
A Golgi rhomboid protease Rbd2 recruits Cdc48 to cleave yeast SREBP.EMBO J. 2016 Nov 2;35(21):2332-2349. doi: 10.15252/embj.201693923. Epub 2016 Sep 21. EMBO J. 2016. PMID: 27655872 Free PMC article.
-
Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase.J Biol Chem. 2012 Jan 2;287(1):672-681. doi: 10.1074/jbc.M111.317370. Epub 2011 Nov 15. J Biol Chem. 2012. PMID: 22086920 Free PMC article.
-
Structural requirements for sterol regulatory element-binding protein (SREBP) cleavage in fission yeast.J Biol Chem. 2013 Jul 12;288(28):20351-60. doi: 10.1074/jbc.M113.482224. Epub 2013 May 31. J Biol Chem. 2013. PMID: 23729666 Free PMC article.
-
Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins.World J Gastroenterol. 2004 Nov 1;10(21):3081-7. doi: 10.3748/wjg.v10.i21.3081. World J Gastroenterol. 2004. PMID: 15457548 Free PMC article. Review.
-
Cdc48-Ufd1-Npl4: stuck in the middle with Ub.Curr Biol. 2002 May 14;12(10):R366-71. doi: 10.1016/s0960-9822(02)00862-x. Curr Biol. 2002. PMID: 12015140 Review.
Cited by
-
The role of rhomboid superfamily members in protein homeostasis: Mechanistic insight and physiological implications.Biochim Biophys Acta Mol Cell Res. 2020 Oct;1867(10):118793. doi: 10.1016/j.bbamcr.2020.118793. Epub 2020 Jul 6. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 32645330 Free PMC article. Review.
-
Sorting of a multi-subunit ubiquitin ligase complex in the endolysosome system.Elife. 2018 Jan 22;7:e33116. doi: 10.7554/eLife.33116. Elife. 2018. PMID: 29355480 Free PMC article.
-
AAA-ATPase valosin-containing protein binds the transcription factor SREBP1 and promotes its proteolytic activation by rhomboid protease RHBDL4.J Biol Chem. 2022 Jun;298(6):101936. doi: 10.1016/j.jbc.2022.101936. Epub 2022 Apr 14. J Biol Chem. 2022. PMID: 35430252 Free PMC article.
-
The Dsc complex and its role in Golgi quality control.Biochem Soc Trans. 2024 Oct 30;52(5):2023-2034. doi: 10.1042/BST20230375. Biochem Soc Trans. 2024. PMID: 39324639 Free PMC article. Review.
-
AMPKα Subunit Ssp2 and Glycogen Synthase Kinases Gsk3/Gsk31 are involved in regulation of sterol regulatory element-binding protein (SREBP) activity in fission yeast.PLoS One. 2020 Feb 13;15(2):e0228845. doi: 10.1371/journal.pone.0228845. eCollection 2020. PLoS One. 2020. PMID: 32053662 Free PMC article.
References
-
- Hughes A. L., Todd B. L., and Espenshade P. J. (2005) SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120, 831–842 - PubMed
-
- Stewart E. V., Lloyd S. J., Burg J. S., Nwosu C. C., Lintner R. E., Daza R., Russ C., Ponchner K., Nusbaum C., and Espenshade P. J. (2012) Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J. Biol. Chem. 287, 672–681 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

