Proteolytic cleavage by the inner membrane peptidase (IMP) complex or Oct1 peptidase controls the localization of the yeast peroxiredoxin Prx1 to distinct mitochondrial compartments
- PMID: 28821623
- PMCID: PMC5641892
- DOI: 10.1074/jbc.M117.788588
Proteolytic cleavage by the inner membrane peptidase (IMP) complex or Oct1 peptidase controls the localization of the yeast peroxiredoxin Prx1 to distinct mitochondrial compartments
Abstract
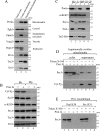
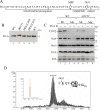
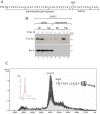
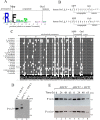
Yeast Prx1 is a mitochondrial 1-Cys peroxiredoxin that catalyzes the reduction of endogenously generated H2O2 Prx1 is synthesized on cytosolic ribosomes as a preprotein with a cleavable N-terminal presequence that is the mitochondrial targeting signal, but the mechanisms underlying Prx1 distribution to distinct mitochondrial subcompartments are unknown. Here, we provide direct evidence of the following dual mitochondrial localization of Prx1: a soluble form in the intermembrane space and a form in the matrix weakly associated with the inner mitochondrial membrane. We show that Prx1 sorting into the intermembrane space likely involves the release of the protein precursor within the lipid bilayer of the inner membrane, followed by cleavage by the inner membrane peptidase. We also found that during its import into the matrix compartment, Prx1 is sequentially cleaved by mitochondrial processing peptidase and then by octapeptidyl aminopeptidase 1 (Oct1). Oct1 cleaved eight amino acid residues from the N-terminal region of Prx1 inside the matrix, without interfering with its peroxidase activity in vitro Remarkably, the processing of peroxiredoxin (Prx) proteins by Oct1 appears to be an evolutionarily conserved process because yeast Oct1 could cleave the human mitochondrial peroxiredoxin Prx3 when expressed in Saccharomyces cerevisiae Altogether, the processing of peroxiredoxins by Imp2 or Oct1 likely represents systems that control the localization of Prxs into distinct compartments and thereby contribute to various mitochondrial redox processes.
Keywords: IMP complex; hydrogen peroxide; inner membrane peptidase complex; mitochondria; mitochondrial processing peptidase (MPP); octapeptidylpeptidase 1, Oct1; oxidative stress; peroxiredoxin; protein import; protein sorting; yeast.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Herrmann J. M., and Riemer J. (2010) The intermembrane space of mitochondria. Antioxid. Redox Signal. 13, 1341–1358 - PubMed
-
- Ryan M. T., and Hoogenraad N. J. (2007) Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76, 701–722 - PubMed
-
- Harbauer A. B., Zahedi R. P., Sickmann A., Pfanner N., and Meisinger C. (2014) The protein import machinery of mitochondria–A regulatory hub in metabolism, stress, and disease. Cell Metab. 19, 357–372 - PubMed
-
- Kowaltowski A. J., de Souza-Pinto N. C., Castilho R. F., and Vercesi A. E. (2009) Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 47, 333–343 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

