Structural and developmental expression of Ss-riok-2, an RIO protein kinase encoding gene of Strongyloides stercoralis
- PMID: 28821723
- PMCID: PMC5562798
- DOI: 10.1038/s41598-017-07991-2
Structural and developmental expression of Ss-riok-2, an RIO protein kinase encoding gene of Strongyloides stercoralis
Abstract
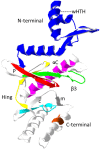
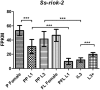
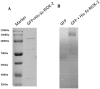
RIO kinases are essential atypical protein kinases in diverse prokaryotic and eukaryotic organisms, playing significant roles in yeast and humans. However, little is known about their functions in parasitic nematodes. In the present study, we have isolated and characterized the full-length cDNA, gDNA and a putative promoter of a RIOK-2 protein kinase (Ss-RIOK-2) encoding gene (Ss-riok-2) from Strongyloides stercoralis, a medically important parasitic nematode (Order Rhabditida). A three-dimensional structure (3D) model of Ss-RIOK-2 was generated using the Chaetomium thermophilum RIOK-2 protein kinase (Ct-RIOK-2) crystal structure 4GYG as a template. A docking study revealed some critical sites for ATP binding and metal binding. The putative promoter of Ss-riok-2 contains a number of conserved elements. RNAseq analysis revealed the highest levels of the Ss-riok-2 transcript in free-living females and parasitic females. To identify anatomical patterns of Ss-riok-2 expression in S. stercoralis, we observed expression patterns of a transgene construct encoding green fluorescent protein under the Ss-riok-2 promoter in post free-living S. stercoralis. Expression driven by this promoter predominated in intestinal cells. This study demonstrates significant advancement in molecular and cellular biological study of S. stercoralis and of parasitic nematodes generally, and provides a foundation for further functional genomic studies.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


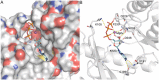


Similar articles
-
Exploring features and function of Ss-riok-3, an enigmatic kinase gene from Strongyloides stercoralis.Parasit Vectors. 2014 Dec 5;7:561. doi: 10.1186/s13071-014-0561-z. Parasit Vectors. 2014. PMID: 25477034 Free PMC article.
-
Functional genomic exploration reveals that Ss-RIOK-1 is essential for the development and survival of Strongyloides stercoralis larvae.Int J Parasitol. 2017 Dec;47(14):933-940. doi: 10.1016/j.ijpara.2017.06.005. Epub 2017 Aug 3. Int J Parasitol. 2017. PMID: 28780152 Free PMC article.
-
Toward understanding the functional role of Ss-RIOK-1, a RIO protein kinase-encoding gene of Strongyloides stercoralis.PLoS Negl Trop Dis. 2014 Aug 7;8(8):e3062. doi: 10.1371/journal.pntd.0003062. eCollection 2014 Aug. PLoS Negl Trop Dis. 2014. PMID: 25101874 Free PMC article.
-
Atypical (RIO) protein kinases from Haemonchus contortus--promise as new targets for nematocidal drugs.Biotechnol Adv. 2011 May-Jun;29(3):338-50. doi: 10.1016/j.biotechadv.2011.01.006. Epub 2011 Jan 22. Biotechnol Adv. 2011. PMID: 21262337 Review.
-
[Strongyloides stercoralis (Bavay, 1876) Stiles et Hassall, 1902 (Nematoa). Part IV. Life cyclec].Wiad Parazytol. 1999;45(1):13-27. Wiad Parazytol. 1999. PMID: 16883713 Review. Polish.
Cited by
-
CRISPR/Cas9 Mutagenesis and Expression of Dominant Mutant Transgenes as Functional Genomic Approaches in Parasitic Nematodes.Front Genet. 2019 Jul 16;10:656. doi: 10.3389/fgene.2019.00656. eCollection 2019. Front Genet. 2019. PMID: 31379923 Free PMC article. Review.
-
Advances in the Molecular and Cellular Biology of Strongyloides spp.Curr Trop Med Rep. 2019 Dec;6(4):161-178. doi: 10.1007/s40475-019-00186-x. Epub 2019 Sep 13. Curr Trop Med Rep. 2019. PMID: 31929961 Free PMC article.
-
A standard workflow for community-driven manual curation of Strongyloides genome annotations.Philos Trans R Soc Lond B Biol Sci. 2024 Jan 15;379(1894):20220443. doi: 10.1098/rstb.2022.0443. Epub 2023 Nov 27. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38008112 Free PMC article.
-
Domain definition and preliminary functional exploration of the endonuclease NOBP-1 in Strongyloides stercoralis.Parasit Vectors. 2023 Nov 3;16(1):399. doi: 10.1186/s13071-023-05940-9. Parasit Vectors. 2023. PMID: 37924155 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

