The inhibitory mechanism of Hal3 on the yeast Ppz1 phosphatase: A mutagenesis analysis
- PMID: 28821821
- PMCID: PMC5562863
- DOI: 10.1038/s41598-017-09360-5
The inhibitory mechanism of Hal3 on the yeast Ppz1 phosphatase: A mutagenesis analysis
Abstract
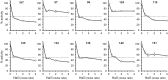
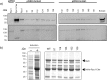
The Ser/Thr protein phosphatase (PPase) Ppz1 is an enzyme related to the ubiquitous type-1 PPases (PP1c) but found only in fungi. It is regulated by an inhibitory subunit, Hal3, which binds to its catalytic domain. Overexpression of Ppz1 is highly toxic for yeast cells, so its de-regulation has been proposed as a target for novel antifungal therapies. While modulation of PP1c by its many regulatory subunits has been extensively characterized, the manner by which Hal3 controls Ppz1 remains unknown. We have used error-prone PCR mutagenesis to construct a library of Ppz1 variants and developed a functional assay to identify mutations affecting the binding or/and the inhibitory capacity of Hal3. We have characterized diverse Ppz1 mutated versions in vivo and in vitro and found that, although they were clearly refractory to Hal3 inhibition, none of them exhibited significant reduction in Hal3 binding. Mapping the mutations strengthened the notion that Hal3 does not interact with Ppz1 through its RVxF-like motif (found in most PP1c regulators). In contrast, the most relevant mutations mapped to a conserved α-helix region used by mammalian Inhibitor-2 to regulate PP1c. Therefore, modulation of PP1c and Ppz1 by their subunits likely differs, but could share some structural features.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Posas F, Casamayor A, Morral N, Arino J. Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino-terminal region. J. Biol. Chem. 1992;267:11734–11740. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

