Expression Profiling of the MAP Kinase Phosphatase Family Reveals a Role for DUSP1 in the Glioblastoma Stem Cell Niche
- PMID: 28822081
- PMCID: PMC5750201
- DOI: 10.1007/s12307-017-0197-6
Expression Profiling of the MAP Kinase Phosphatase Family Reveals a Role for DUSP1 in the Glioblastoma Stem Cell Niche
Abstract
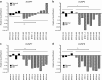
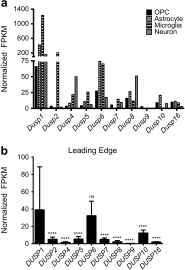
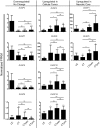
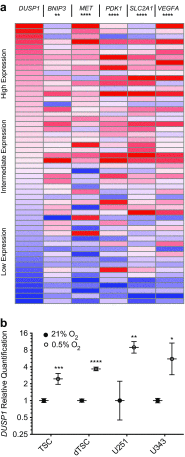
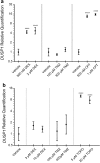
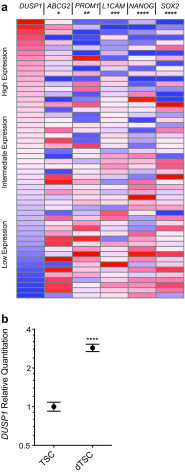
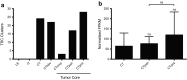
The dual specificity phosphatases (DUSPs) constitute a family of stress-induced enzymes that provide feedback inhibition on mitogen-activated protein kinases (MAPKs) critical in key aspects of oncogenic signaling. While described in other tumor types, the landscape of DUSP mRNA expression in glioblastoma (GB) remains largely unexplored. Interrogation of the REpository for Molecular BRAin Neoplasia DaTa (REMBRANDT) revealed induction (DUSP4, DUSP6), repression (DUSP2, DUSP7-9), or mixed (DUSP1, DUSP5, DUSP10, DUSP15) DUSP transcription of select DUSPs in bulk tumor specimens. To resolve features specific to the tumor microenvironment, we searched the Ivy Glioblastoma Atlas Project (Ivy GAP) repository, which highlight DUSP1, DUSP5, and DUSP6 as the predominant family members induced within pseudopalisading and perinecrotic regions. The inducibility of DUSP1 in response to hypoxia, dexamethasone, or the chemotherapeutic agent camptothecin was confirmed in GB cell lines and tumor-derived stem cells (TSCs). Moreover, we show that loss of DUSP1 expression is a characteristic of TSCs and correlates with expression of tumor stem cell markers in situ (ABCG2, PROM1, L1CAM, NANOG, SOX2). This work reveals a dynamic pattern of DUSP expression within the tumor microenvironment that reflects the cumulative effects of factors including regional ischemia, chemotherapeutic exposure among others. Moreover, our observation regarding DUSP1 dysregulation within the stem cell niche argue for its importance in the survival and proliferation of this therapeutically resistant population.
Keywords: Camptothecin; DUSP1; Dexamethasone; Glioblastoma multiforme; Hypoxia; Mitogen-activated protein kinases; Temozolomide; Tumor stem cell.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

