Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders
- PMID: 28824374
- PMCID: PMC5540894
- DOI: 10.3389/fnmol.2017.00243
Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders
Abstract
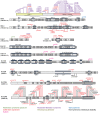
The ATP-dependent BRG1/BRM associated factor (BAF) chromatin remodeling complexes are crucial in regulating gene expression by controlling chromatin dynamics. Over the last decade, it has become increasingly clear that during neural development in mammals, distinct ontogenetic stage-specific BAF complexes derived from combinatorial assembly of their subunits are formed in neural progenitors and post-mitotic neural cells. Proper functioning of the BAF complexes plays critical roles in neural development, including the establishment and maintenance of neural fates and functionality. Indeed, recent human exome sequencing and genome-wide association studies have revealed that mutations in BAF complex subunits are linked to neurodevelopmental disorders such as Coffin-Siris syndrome, Nicolaides-Baraitser syndrome, Kleefstra's syndrome spectrum, Hirschsprung's disease, autism spectrum disorder, and schizophrenia. In this review, we focus on the latest insights into the functions of BAF complexes during neural development and the plausible mechanistic basis of how mutations in known BAF subunits are associated with certain neurodevelopmental disorders.
Keywords: BAF (mSWI/SNF) complex; chromatin remodeling; epigenetic regulation; neural development; neurodevelopmental disorder.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

