Pan-genome Analyses of the Species Salmonella enterica, and Identification of Genomic Markers Predictive for Species, Subspecies, and Serovar
- PMID: 28824552
- PMCID: PMC5534482
- DOI: 10.3389/fmicb.2017.01345
Pan-genome Analyses of the Species Salmonella enterica, and Identification of Genomic Markers Predictive for Species, Subspecies, and Serovar
Abstract
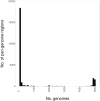
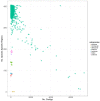
Food safety is a global concern, with upward of 2.2 million deaths due to enteric disease every year. Current whole-genome sequencing platforms allow routine sequencing of enteric pathogens for surveillance, and during outbreaks; however, a remaining challenge is the identification of genomic markers that are predictive of strain groups that pose the most significant health threats to humans, or that can persist in specific environments. We have previously developed the software program Panseq, which identifies the pan-genome among a group of sequences, and the SuperPhy platform, which utilizes this pan-genome information to identify biomarkers that are predictive of groups of bacterial strains. In this study, we examined the pan-genome of 4893 genomes of Salmonella enterica, an enteric pathogen responsible for the loss of more disability adjusted life years than any other enteric pathogen. We identified a pan-genome of 25.3 Mbp, a strict core of 1.5 Mbp present in all genomes, and a conserved core of 3.2 Mbp found in at least 96% of these genomes. We also identified 404 genomic regions of 1000 bp that were specific to the species S. enterica. These species-specific regions were found to encode mostly hypothetical proteins, effectors, and other proteins related to virulence. For each of the six S. enterica subspecies, markers unique to each were identified. No serovar had pan-genome regions that were present in all of its genomes and absent in all other serovars; however, each serovar did have genomic regions that were universally present among all constituent members, and statistically predictive of the serovar. The phylogeny based on SNPs within the conserved core genome was found to be highly concordant to that produced by a phylogeny using the presence/absence of 1000 bp regions of the entire pan-genome. Future studies could use these predictive regions as components of a vaccine to prevent salmonellosis, as well as in simple and rapid diagnostic tests for both in silico and wet-lab applications, with uses ranging from food safety to public health. Lastly, the tools and methods described in this study could be applied as a pan-genomics framework to other population genomic studies seeking to identify markers for other bacterial species and their sub-groups.
Keywords: Salmonella; food safety; genomics; pan-genome; predictive markers.
Figures





Similar articles
-
Identification of Salmonella enterica species- and subgroup-specific genomic regions using Panseq 2.0.Infect Genet Evol. 2011 Dec;11(8):2151-61. doi: 10.1016/j.meegid.2011.09.021. Epub 2011 Oct 1. Infect Genet Evol. 2011. PMID: 22001825
-
Genomic characterization of endemic Salmonella enterica serovar Typhimurium and Salmonella enterica serovar I 4,[5],12:i:- isolated in Malaysia.Infect Genet Evol. 2018 Aug;62:109-121. doi: 10.1016/j.meegid.2018.04.027. Epub 2018 Apr 21. Infect Genet Evol. 2018. PMID: 29684710
-
Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions.BMC Bioinformatics. 2010 Sep 15;11:461. doi: 10.1186/1471-2105-11-461. BMC Bioinformatics. 2010. PMID: 20843356 Free PMC article.
-
Pan-genome: setting a new standard for high-quality reference genomes.Yi Chuan. 2021 Nov 20;43(11):1023-1037. doi: 10.16288/j.yczz.21-214. Yi Chuan. 2021. PMID: 34815206 Review.
-
Plant pan-genomics and its applications.Mol Plant. 2023 Jan 2;16(1):168-186. doi: 10.1016/j.molp.2022.12.009. Epub 2022 Dec 15. Mol Plant. 2023. PMID: 36523157 Review.
Cited by
-
Performance and Accuracy of Four Open-Source Tools for In Silico Serotyping of Salmonella spp. Based on Whole-Genome Short-Read Sequencing Data.Appl Environ Microbiol. 2020 Feb 18;86(5):e02265-19. doi: 10.1128/AEM.02265-19. Print 2020 Feb 18. Appl Environ Microbiol. 2020. PMID: 31862714 Free PMC article.
-
Large-Scale Genomics Reveals the Genetic Characteristics of Seven Species and Importance of Phylogenetic Distance for Estimating Pan-Genome Size.Front Microbiol. 2019 Apr 24;10:834. doi: 10.3389/fmicb.2019.00834. eCollection 2019. Front Microbiol. 2019. PMID: 31068915 Free PMC article.
-
Comparative Genomic Analyses and CRISPR-Cas Characterization of Cutibacterium acnes Provide Insights Into Genetic Diversity and Typing Applications.Front Microbiol. 2021 Nov 3;12:758749. doi: 10.3389/fmicb.2021.758749. eCollection 2021. Front Microbiol. 2021. PMID: 34803983 Free PMC article.
-
Mobile genetic elements define the non-random structure of the Salmonella enterica serovar Typhi pangenome.mSystems. 2024 Aug 20;9(8):e0036524. doi: 10.1128/msystems.00365-24. Epub 2024 Jul 26. mSystems. 2024. PMID: 39058093 Free PMC article.
-
Recent emergence of cephalosporin-resistant Salmonella Typhi in India due to the endemic clone acquiring IncFIB(K) plasmid encoding blaCTX-M-15 gene.Microbiol Spectr. 2025 Apr 10;13(5):e0087524. doi: 10.1128/spectrum.00875-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40208005 Free PMC article.
References
-
- Aanensen D. M., Feil E. J., Holden M. T. G., Dordel J., Yeats C. A., Fedosejev A., et al. (2016). Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 7:e00444–16 10.1128/mBio.00444-16 - DOI - PMC - PubMed
-
- Babenko D., Azizov I., Toleman M. (2016). wgMLST as a standardized tool for assessing the quality of genome assembly data. Int. J. Infect. Dis. 45:329 10.1016/j.ijid.2016.02.714 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

