Diaporthe is paraphyletic
- PMID: 28824846
- PMCID: PMC5493532
- DOI: 10.5598/imafungus.2017.08.01.11
Diaporthe is paraphyletic
Abstract
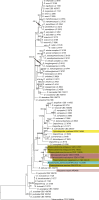
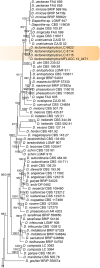
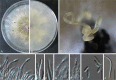
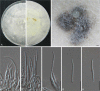
Previous studies have shown that our understanding of species diversity within Diaporthe (syn. Phomopsis) is limited. In this study, 49 strains obtained from different countries were subjected to DNA sequence analysis. Based on these results, eight new species names are introduced for lineages represented by multiple strains and distinct morphology. Twelve Phomopsis species previously described from China were subjected to DNA sequence analysis, and confirmed to belong to Diaporthe. The genus Diaporthe is shown to be paraphyletic based on multi-locus (LSU, ITS and TEF1) phylogenetic analysis. Several morphologically distinct genera, namely Mazzantia, Ophiodiaporthe, Pustulomyces, Phaeocytostroma, and Stenocarpella, are embedded within Diaporthe s. lat., indicating divergent morphological evolution. However, splitting Diaporthe into many smaller genera to achieve monophyly is still premature, and further collections and phylogenetic datasets need to be obtained to address this situation.
Keywords: Ascomycota; Diaporthales; Phomopsis; phylogeny; taxonomy.
Figures







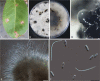

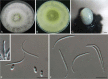

References
-
- Annesi T, Luongo L, Vitale S, Galli M, Belisario A. (2015) Characterization and pathogenicity of Phomopsis theicola anamorph of Diaporthe foeniculina causing stem and shoot cankers on sweet chestnut in Italy. Journal of Phytopathology 164: 412–416.
-
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556.
-
- Castlebury LA, Rossman AY, Jaklitsch WJ, Vasilyeva L. (2002) A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia 94: 1017–1031. - PubMed
-
- Chang CQ, Cheng YH, Xiang MM, Jiang ZD. (2005) New species of Phomopsis on woody plants in Fujian Province. Mycosystema 24: 6–11.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
