Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making
- PMID: 28825054
- PMCID: PMC5558263
- DOI: 10.1200/PO.17.00029
Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making
Abstract
Purpose: A long natural history and a predominant osseous pattern of metastatic spread are impediments to the adoption of precision medicine in patients with prostate cancer. To establish the feasibility of clinical genomic profiling in the disease, we performed targeted deep sequencing of tumor and normal DNA from patients with locoregional, metastatic non-castrate, and metastatic castration-resistant prostate cancer (CRPC).
Methods: Patients consented to genomic analysis of their tumor and germline DNA. A hybridization capture-based clinical assay was employed to identify single nucleotide variations, small insertions and deletions, copy number alterations and structural rearrangements in over 300 cancer-related genes in tumors and matched normal blood.
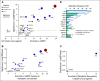
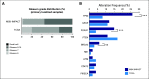
Results: We successfully sequenced 504 tumors from 451 patients with prostate cancer. Potentially actionable alterations were identified in DNA damage repair (DDR), PI3K, and MAP kinase pathways. 27% of patients harbored a germline or a somatic alteration in a DDR gene that may predict for response to PARP inhibition. Profiling of matched tumors from individual patients revealed that somatic TP53 and BRCA2 alterations arose early in tumors from patients who eventually developed metastatic disease. In contrast, comparative analysis across disease states revealed that APC alterations were enriched in metastatic tumors, while ATM alterations were specifically enriched in CRPC.
Conclusion: Through genomic profiling of prostate tumors representing the disease clinical spectrum, we identified a high frequency of potentially actionable alterations and possible drivers of disease initiation, metastasis and castration-resistance. Our findings support the routine use of tumor and germline DNA profiling for patients with advanced prostate cancer, for the purpose of guiding enrollment in targeted clinical trials and counseling families at increased risk of malignancy.
Conflict of interest statement
Authors’ disclosures of potential conflicts of interest No conflicts of interest to declare.
Figures
















References
-
- Scher HI, Heller G. Clinical states in prostate cancer: Toward a dynamic model of disease progression. Urology. 2000;55:323–327. - PubMed
-
- National Comprehensive Cancer Network Prostate cancer (version 1.2015) https://www.nccn.org/patients/guidelines/prostate/files/assets/common/do... - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

