Recapitulating in vivo-like plasticity of glioma cell invasion along blood vessels and in astrocyte-rich stroma
- PMID: 28825130
- PMCID: PMC5602046
- DOI: 10.1007/s00418-017-1604-2
Recapitulating in vivo-like plasticity of glioma cell invasion along blood vessels and in astrocyte-rich stroma
Abstract
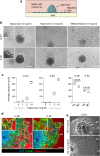
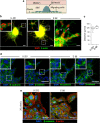
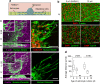
Diffuse invasion of glioma cells into the brain parenchyma leads to nonresectable brain tumors and poor prognosis of glioma disease. In vivo, glioma cells can adopt a range of invasion strategies and routes, by moving as single cells, collective strands and multicellular networks along perivascular, perineuronal and interstitial guidance cues. Current in vitro assays to probe glioma cell invasion, however, are limited in recapitulating the modes and adaptability of glioma invasion observed in brain parenchyma, including collective behaviours. To mimic in vivo-like glioma cell invasion in vitro, we here applied three tissue-inspired 3D environments combining multicellular glioma spheroids and reconstituted microanatomic features of vascular and interstitial brain structures. Radial migration from multicellular glioma spheroids of human cell lines and patient-derived xenograft cells was monitored using (1) reconstituted basement membrane/hyaluronan interfaces representing the space along brain vessels; (2) 3D scaffolds generated by multi-layered mouse astrocytes to reflect brain interstitium; and (3) freshly isolated mouse brain slice culture ex vivo. The invasion patterns in vitro were validated using histological analysis of brain sections from glioblastoma patients and glioma xenografts infiltrating the mouse brain. Each 3D assay recapitulated distinct aspects of major glioma invasion patterns identified in mouse xenografts and patient brain samples, including individually migrating cells, collective strands extending along blood vessels, and multicellular networks of interconnected glioma cells infiltrating the neuropil. In conjunction, these organotypic assays enable a range of invasion modes used by glioma cells and will be applicable for mechanistic analysis and targeting of glioma cell dissemination.
Keywords: Astrocyte scaffolds; Glioma invasion; Multicellular networks; Organotypic culture; Perivascular invasion.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Albini A, Iwamoto Y, Kleinman HK, et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

