Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis
- PMID: 28825595
- PMCID: PMC5669571
- DOI: 10.1172/JCI94292
Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis
Abstract
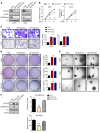
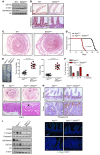
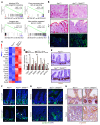
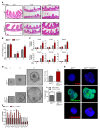
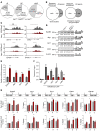
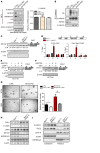
The histone H3K36 methyltransferase SETD2 is frequently mutated or deleted in a variety of human tumors. Nevertheless, the role of SETD2 loss in oncogenesis remains largely undefined. Here, we found that SETD2 counteracts Wnt signaling and its inactivation promotes intestinal tumorigenesis in mouse models of colorectal cancer (CRC). SETD2 was not required for intestinal homeostasis under steady state; however, upon irradiation, genetic inactivation of Setd2 in mouse intestinal epithelium facilitated the self-renewal of intestinal stem/progenitor cells as well as tissue regeneration. Furthermore, depletion of SETD2 enhanced the susceptibility to tumorigenesis in the context of dysregulated Wnt signaling. Mechanistic characterizations indicated that SETD2 downregulation affects the alternative splicing of a subset of genes implicated in tumorigenesis. Importantly, we uncovered that SETD2 ablation reduces intron retention of dishevelled segment polarity protein 2 (DVL2) pre-mRNA, which would otherwise be degraded by nonsense-mediated decay, thereby augmenting Wnt signaling. The signaling cascades mediated by SETD2 were further substantiated by a CRC patient cohort analysis. Together, our studies highlight SETD2 as an integral regulator of Wnt signaling through epigenetic regulation of RNA processing during tissue regeneration and tumorigenesis.
Conflict of interest statement
Figures


Comment in
-
Epigenetics at the base of alternative splicing changes that promote colorectal cancer.J Clin Invest. 2017 Sep 1;127(9):3281-3283. doi: 10.1172/JCI96497. Epub 2017 Aug 21. J Clin Invest. 2017. PMID: 28825597 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

