Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors
- PMID: 28825697
- PMCID: PMC5633079
- DOI: 10.1038/ncb3597
Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors
Abstract
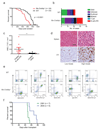
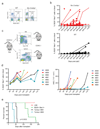
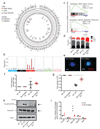
Loss-of-function mutations of cyclic-AMP response element binding protein, binding protein (CREBBP) are prevalent in lymphoid malignancies. However, the tumour suppressor functions of CREBBP remain unclear. We demonstrate that loss of Crebbp in murine haematopoietic stem and progenitor cells (HSPCs) leads to increased development of B-cell lymphomas. This is preceded by accumulation of hyperproliferative lymphoid progenitors with a defective DNA damage response (DDR) due to a failure to acetylate p53. We identify a premalignant lymphoma stem cell population with decreased H3K27ac, which undergoes transcriptional and genetic evolution due to the altered DDR, resulting in lymphomagenesis. Importantly, when Crebbp is lost later in lymphopoiesis, cellular abnormalities are lost and tumour generation is attenuated. We also document that CREBBP mutations may occur in HSPCs from patients with CREBBP-mutated lymphoma. These data suggest that earlier loss of Crebbp is advantageous for lymphoid transformation and inform the cellular origins and subsequent evolution of lymphoid malignancies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
MeSH terms
Substances
Grants and funding
- MR/M008584/1/MRC_/Medical Research Council/United Kingdom
- MR/M010392/1/MRC_/Medical Research Council/United Kingdom
- 13031/CRUK_/Cancer Research UK/United Kingdom
- MR/L019027/1/MRC_/Medical Research Council/United Kingdom
- MR/M008975/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_12009/MRC_/Medical Research Council/United Kingdom
- G1000288/MRC_/Medical Research Council/United Kingdom
- 14-1069/AICR_/Worldwide Cancer Research/United Kingdom
- 15968/CRUK_/Cancer Research UK/United Kingdom
- 647685/ERC_/European Research Council/International
- 21717/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

