SMC complexes differentially compact mitotic chromosomes according to genomic context
- PMID: 28825700
- PMCID: PMC5640152
- DOI: 10.1038/ncb3594
SMC complexes differentially compact mitotic chromosomes according to genomic context
Abstract
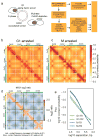
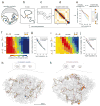
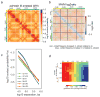
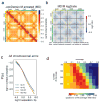
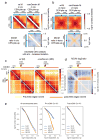
Structural maintenance of chromosomes (SMC) protein complexes are key determinants of chromosome conformation. Using Hi-C and polymer modelling, we study how cohesin and condensin, two deeply conserved SMC complexes, organize chromosomes in the budding yeast Saccharomyces cerevisiae. The canonical role of cohesin is to co-align sister chromatids, while condensin generally compacts mitotic chromosomes. We find strikingly different roles for the two complexes in budding yeast mitosis. First, cohesin is responsible for compacting mitotic chromosome arms, independently of sister chromatid cohesion. Polymer simulations demonstrate that this role can be fully accounted for through cis-looping of chromatin. Second, condensin is generally dispensable for compaction along chromosome arms. Instead, it plays a targeted role compacting the rDNA proximal regions and promoting resolution of peri-centromeric regions. Our results argue that the conserved mechanism of SMC complexes is to form chromatin loops and that distinct SMC-dependent looping activities are selectively deployed to appropriately compact chromosomes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Chromosome biology: Different turfs for cohesin and condensin.Nat Rev Mol Cell Biol. 2017 Oct;18(10):592-593. doi: 10.1038/nrm.2017.90. Epub 2017 Aug 23. Nat Rev Mol Cell Biol. 2017. PMID: 28831204 No abstract available.
-
Chromosome biology: Different turfs for cohesin and condensin.Nat Rev Genet. 2017 Oct;18(10):578-579. doi: 10.1038/nrg.2017.71. Epub 2017 Aug 30. Nat Rev Genet. 2017. PMID: 28852222 No abstract available.
-
Chromosome structure dynamics during the cell cycle: a structure to fit every phase.EMBO J. 2017 Sep 15;36(18):2661-2663. doi: 10.15252/embj.201798014. Epub 2017 Sep 4. EMBO J. 2017. PMID: 28871059 Free PMC article.
References
-
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

