Condensin-mediated remodeling of the mitotic chromatin landscape in fission yeast
- PMID: 28825727
- PMCID: PMC5621628
- DOI: 10.1038/ng.3938
Condensin-mediated remodeling of the mitotic chromatin landscape in fission yeast
Abstract
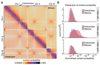
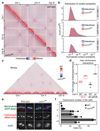
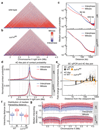
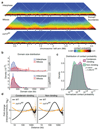
The eukaryotic genome consists of DNA molecules far longer than the cells that contain them. They reach their greatest compaction during chromosome condensation in mitosis. This process is aided by condensin, a structural maintenance of chromosomes (SMC) family member. The spatial organization of mitotic chromosomes and how condensin shapes chromatin architecture are not yet fully understood. Here we use chromosome conformation capture (Hi-C) to study mitotic chromosome condensation in the fission yeast Schizosaccharomyces pombe. This showed that the interphase landscape characterized by small chromatin domains is replaced by fewer but larger domains in mitosis. Condensin achieves this by setting up longer-range, intrachromosomal DNA interactions, which compact and individualize chromosomes. At the same time, local chromatin contacts are constrained by condensin, with profound implications for local chromatin function during mitosis. Our results highlight condensin as a major determinant that changes the chromatin landscape as cells prepare their genomes for cell division.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Comment in
-
Chromosome biology: Different turfs for cohesin and condensin.Nat Rev Genet. 2017 Oct;18(10):578-579. doi: 10.1038/nrg.2017.71. Epub 2017 Aug 30. Nat Rev Genet. 2017. PMID: 28852222 No abstract available.
-
Chromosome structure dynamics during the cell cycle: a structure to fit every phase.EMBO J. 2017 Sep 15;36(18):2661-2663. doi: 10.15252/embj.201798014. Epub 2017 Sep 4. EMBO J. 2017. PMID: 28871059 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

