Extensive alternative splicing transitions during postnatal skeletal muscle development are required for calcium handling functions
- PMID: 28826478
- PMCID: PMC5577920
- DOI: 10.7554/eLife.27192
Extensive alternative splicing transitions during postnatal skeletal muscle development are required for calcium handling functions
Abstract
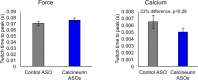
Postnatal development of skeletal muscle is a highly dynamic period of tissue remodeling. Here, we used RNA-seq to identify transcriptome changes from late embryonic to adult mouse muscle and demonstrate that alternative splicing developmental transitions impact muscle physiology. The first 2 weeks after birth are particularly dynamic for differential gene expression and alternative splicing transitions, and calcium-handling functions are significantly enriched among genes that undergo alternative splicing. We focused on the postnatal splicing transitions of the three calcineurin A genes, calcium-dependent phosphatases that regulate multiple aspects of muscle biology. Redirected splicing of calcineurin A to the fetal isoforms in adult muscle and in differentiated C2C12 slows the timing of muscle relaxation, promotes nuclear localization of calcineurin target Nfatc3, and/or affects expression of Nfatc transcription targets. The results demonstrate a previously unknown specificity of calcineurin isoforms as well as the broader impact of alternative splicing during muscle postnatal development.
Keywords: Nfat; alternative splicing; calcineurin; cell biology; developmental biology; none; postnatal development; skeletal muscle; stem cells.
Conflict of interest statement
No competing interests declared.
Figures










Similar articles
-
Misregulation of calcium-handling proteins promotes hyperactivation of calcineurin-NFAT signaling in skeletal muscle of DM1 mice.Hum Mol Genet. 2017 Jun 15;26(12):2192-2206. doi: 10.1093/hmg/ddx109. Hum Mol Genet. 2017. PMID: 28369518
-
Alternative Splicing of Four Trafficking Genes Regulates Myofiber Structure and Skeletal Muscle Physiology.Cell Rep. 2016 Nov 15;17(8):1923-1933. doi: 10.1016/j.celrep.2016.10.072. Cell Rep. 2016. PMID: 27851958 Free PMC article.
-
Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway.J Cell Physiol. 2007 Apr;211(1):138-48. doi: 10.1002/jcp.20916. J Cell Physiol. 2007. PMID: 17111365
-
Parallel mechanisms for resting nucleo-cytoplasmic shuttling and activity dependent translocation provide dual control of transcriptional regulators HDAC and NFAT in skeletal muscle fiber type plasticity.J Muscle Res Cell Motil. 2006;27(5-7):405-11. doi: 10.1007/s10974-006-9080-7. Epub 2006 Jul 28. J Muscle Res Cell Motil. 2006. PMID: 16874450 Review.
-
Remodeling muscles with calcineurin.Bioessays. 2000 Jun;22(6):510-9. doi: 10.1002/(SICI)1521-1878(200006)22:6<510::AID-BIES4>3.0.CO;2-1. Bioessays. 2000. PMID: 10842305 Review.
Cited by
-
SRSF2 is a key player in orchestrating the directional migration and differentiation of MyoD progenitors during skeletal muscle development.Elife. 2024 Sep 9;13:RP98175. doi: 10.7554/eLife.98175. Elife. 2024. PMID: 39248331 Free PMC article.
-
Stretching muscle cells induces transcriptional and splicing transitions and changes in SR proteins.Commun Biol. 2022 Sep 19;5(1):987. doi: 10.1038/s42003-022-03915-7. Commun Biol. 2022. PMID: 36123433 Free PMC article.
-
miR-1/206 downregulates splicing factor Srsf9 to promote C2C12 differentiation.Skelet Muscle. 2019 Dec 2;9(1):31. doi: 10.1186/s13395-019-0211-4. Skelet Muscle. 2019. PMID: 31791406 Free PMC article.
-
Reciprocal regulation of hnRNP C and CELF2 through translation and transcription tunes splicing activity in T cells.Nucleic Acids Res. 2020 Jun 4;48(10):5710-5719. doi: 10.1093/nar/gkaa295. Nucleic Acids Res. 2020. PMID: 32338744 Free PMC article.
-
Dynamic Changes in the Global Transcriptome of Postnatal Skeletal Muscle in Different Sheep.Genes (Basel). 2023 Jun 20;14(6):1298. doi: 10.3390/genes14061298. Genes (Basel). 2023. PMID: 37372481 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

