Munc18a clusters SNARE-bearing liposomes prior to trans-SNARE zippering
- PMID: 28827281
- PMCID: PMC5651683
- DOI: 10.1042/BCJ20170494
Munc18a clusters SNARE-bearing liposomes prior to trans-SNARE zippering
Abstract
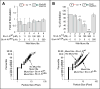
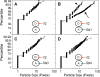
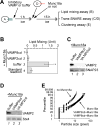
Sec1-Munc18 (SM) proteins co-operate with SNAREs {SNAP [soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein] receptors} to mediate membrane fusion in eukaryotic cells. Studies of Munc18a/Munc18-1/Stxbp1 in neurotransmission suggest that SM proteins accelerate fusion kinetics primarily by activating the partially zippered trans-SNARE complex. However, accumulating evidence has argued for additional roles for SM proteins in earlier steps in the fusion cascade. Here, we investigate the function of Munc18a in reconstituted exocytic reactions mediated by neuronal and non-neuronal SNAREs. We show that Munc18a plays a direct role in promoting proteoliposome clustering, underlying vesicle docking during exocytosis. In the three different fusion reactions examined, Munc18a-dependent clustering requires an intact N-terminal peptide (N-peptide) motif in syntaxin that mediates the binary interaction between syntaxin and Munc18a. Importantly, clustering is preserved under inhibitory conditions that abolish both trans-SNARE complex formation and lipid mixing, indicating that Munc18a promotes membrane clustering in a step that is independent of trans-SNARE zippering and activation.
Keywords: Munc18; SNARE proteins; docking; exocytosis; membrane fusion; reconstitution.
© 2017 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

