Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol
- PMID: 28827342
- PMCID: PMC5604016
- DOI: 10.1073/pnas.1706070114
Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol
Erratum in
-
Correction to Supporting Information for Alvarez et al., Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol.Proc Natl Acad Sci U S A. 2017 Dec 12;114(50):E10852. doi: 10.1073/pnas.1719818115. Epub 2017 Nov 27. Proc Natl Acad Sci U S A. 2017. PMID: 29180409 Free PMC article. No abstract available.
-
Correction for Alvarez et al., Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol.Proc Natl Acad Sci U S A. 2017 Dec 12;114(50):E10849-E10850. doi: 10.1073/pnas.1719819115. Epub 2017 Nov 27. Proc Natl Acad Sci U S A. 2017. PMID: 29180411 Free PMC article. No abstract available.
Abstract
Risk, severity, and outcome of infection depend on the interplay of pathogen virulence and host susceptibility. Systematic identification of genetic susceptibility to infection is being undertaken through genome-wide association studies, but how to expeditiously move from genetic differences to functional mechanisms is unclear. Here, we use genetic association of molecular, cellular, and human disease traits and experimental validation to demonstrate that genetic variation affects expression of VAC14, a phosphoinositide-regulating protein, to influence susceptibility to Salmonella enterica serovar Typhi (S Typhi) infection. Decreased VAC14 expression increased plasma membrane cholesterol, facilitating Salmonella docking and invasion. This increased susceptibility at the cellular level manifests as increased susceptibility to typhoid fever in a Vietnamese population. Furthermore, treating zebrafish with a cholesterol-lowering agent, ezetimibe, reduced susceptibility to S Typhi. Thus, coupling multiple genetic association studies with mechanistic dissection revealed how VAC14 regulates Salmonella invasion and typhoid fever susceptibility and may open doors to new prophylactic/therapeutic approaches.
Keywords: Salmonella pathogenicity island 1; ezetimibe; lymphoblastoid cell line; phosphoinositide; single nucleotide polymorphism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


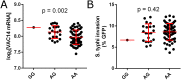


Comment in
-
Genetic variation in VAC14 is associated with bacteremia secondary to diverse pathogens in African children.Proc Natl Acad Sci U S A. 2018 Apr 17;115(16):E3601-E3603. doi: 10.1073/pnas.1802071115. Epub 2018 Mar 27. Proc Natl Acad Sci U S A. 2018. PMID: 29588414 Free PMC article. No abstract available.
-
Reply to Gilchrist et al.: Possible roles for VAC14 in multiple infectious diseases.Proc Natl Acad Sci U S A. 2018 Apr 17;115(16):E3604-E3605. doi: 10.1073/pnas.1803533115. Epub 2018 Mar 27. Proc Natl Acad Sci U S A. 2018. PMID: 29588421 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

