CD146 coordinates brain endothelial cell-pericyte communication for blood-brain barrier development
- PMID: 28827364
- PMCID: PMC5594696
- DOI: 10.1073/pnas.1710848114
CD146 coordinates brain endothelial cell-pericyte communication for blood-brain barrier development
Abstract
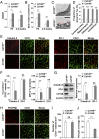
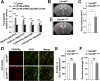
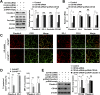
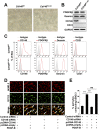
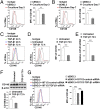
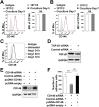
The blood-brain barrier (BBB) establishes a protective interface between the central neuronal system and peripheral blood circulation and is crucial for homeostasis of the CNS. BBB formation starts when the endothelial cells (ECs) invade the CNS and pericytes are recruited to the nascent vessels during embryogenesis. Despite the essential function of pericyte-EC interaction during BBB development, the molecular mechanisms coordinating the pericyte-EC behavior and communication remain incompletely understood. Here, we report a single cell receptor, CD146, that presents dynamic expression patterns in the cerebrovasculature at the stages of BBB induction and maturation, coordinates the interplay of ECs and pericytes, and orchestrates BBB development spatiotemporally. In mouse brain, CD146 is first expressed in the cerebrovascular ECs of immature capillaries without pericyte coverage; with increased coverage of pericytes, CD146 could only be detected in pericytes, but not in cerebrovascular ECs. Specific deletion of Cd146 in mice ECs resulted in reduced brain endothelial claudin-5 expression and BBB breakdown. By analyzing mice with specific deletion of Cd146 in pericytes, which have defects in pericyte coverage and BBB integrity, we demonstrate that CD146 functions as a coreceptor of PDGF receptor-β to mediate pericyte recruitment to cerebrovascular ECs. Moreover, we found that the attached pericytes in turn down-regulate endothelial CD146 by secreting TGF-β1 to promote further BBB maturation. These results reveal that the dynamic expression of CD146 controls the behavior of ECs and pericytes, thereby coordinating the formation of a mature and stable BBB.
Keywords: CD146; PDGFRβ; blood–brain barrier; claudin-5; spatiotemporal expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures












References
-
- Abdelilah-Seyfried S. Claudin-5a in developing zebrafish brain barriers: Another brick in the wall. BioEssays. 2010;32:768–776. - PubMed
-
- Haseloff RF, Dithmer S, Winkler L, Wolburg H, Blasig IE. Transmembrane proteins of the tight junctions at the blood-brain barrier: Structural and functional aspects. Semin Cell Dev Biol. 2015;38:16–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

