Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers
- PMID: 28827559
- PMCID: PMC5566552
- DOI: 10.1038/s41598-017-08273-7
Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers
Abstract
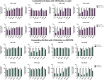
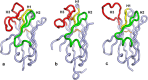
Broadly neutralizing antibodies (bnAbs) against HIV-1 protect from infection and reduce viral load upon therapeutic applications. However no vaccine was able so far to induce bnAbs demanding their expensive biotechnological production. For clinical applications, nanobodies (VHH) derived from heavy chain only antibodies from Camelidae, may be better suited due to their small size, high solubility/stability and extensive homology to human VH3 genes. Here we selected broadly neutralizing nanobodies by phage display after immunization of dromedaries with different soluble trimeric envelope proteins derived from HIV-1 subtype C. We identified 25 distinct VHH families binding trimeric Env, of which 6 neutralized heterologous primary isolates of various HIV-1 subtypes in a standardized in vitro neutralization assay. The complementary neutralization pattern of two selected VHHs in combination covers 19 out of 21 HIV-1 strains from a standardized panel of epidemiologically relevant HIV-1 subtypes. The CD4 binding site was preferentially targeted by the broadly neutralizing VHHs as determined by competition ELISAs and 3D models of VHH-Env complexes derived from negative stain electron microscopy. The nanobodies identified here are excellent candidates for further preclinical/clinical development for prophylactic and therapeutic applications due to their potency and their complementary neutralization patterns covering the majority of epidemiologically relevant HIV-1 subtypes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Geiss Y, Dietrich U. Catch Me If You Can–The Race Between HIV and Neutralizing Antibodies. AIDS Rev. 2015;17:107–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

