Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis
- PMID: 28827734
- PMCID: PMC5566454
- DOI: 10.1038/s41598-017-09335-6
Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis
Abstract
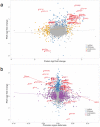
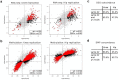
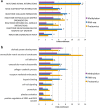
Osteoarthritis (OA) is a common disease characterized by cartilage degeneration and joint remodeling. The underlying molecular changes underpinning disease progression are incompletely understood. We investigated genes and pathways that mark OA progression in isolated primary chondrocytes taken from paired intact versus degraded articular cartilage samples across 38 patients undergoing joint replacement surgery (discovery cohort: 12 knee OA, replication cohorts: 17 knee OA, 9 hip OA patients). We combined genome-wide DNA methylation, RNA sequencing, and quantitative proteomics data. We identified 49 genes differentially regulated between intact and degraded cartilage in at least two -omics levels, 16 of which have not previously been implicated in OA progression. Integrated pathway analysis implicated the involvement of extracellular matrix degradation, collagen catabolism and angiogenesis in disease progression. Using independent replication datasets, we showed that the direction of change is consistent for over 90% of differentially expressed genes and differentially methylated CpG probes. AQP1, COL1A1 and CLEC3B were significantly differentially regulated across all three -omics levels, confirming their differential expression in human disease. Through integration of genome-wide methylation, gene and protein expression data in human primary chondrocytes, we identified consistent molecular players in OA progression that replicated across independent datasets and that have translational potential.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

