An integrated expression atlas of miRNAs and their promoters in human and mouse
- PMID: 28829439
- PMCID: PMC5767576
- DOI: 10.1038/nbt.3947
An integrated expression atlas of miRNAs and their promoters in human and mouse
Abstract
MicroRNAs (miRNAs) are short non-coding RNAs with key roles in cellular regulation. As part of the fifth edition of the Functional Annotation of Mammalian Genome (FANTOM5) project, we created an integrated expression atlas of miRNAs and their promoters by deep-sequencing 492 short RNA (sRNA) libraries, with matching Cap Analysis Gene Expression (CAGE) data, from 396 human and 47 mouse RNA samples. Promoters were identified for 1,357 human and 804 mouse miRNAs and showed strong sequence conservation between species. We also found that primary and mature miRNA expression levels were correlated, allowing us to use the primary miRNA measurements as a proxy for mature miRNA levels in a total of 1,829 human and 1,029 mouse CAGE libraries. We thus provide a broad atlas of miRNA expression and promoters in primary mammalian cells, establishing a foundation for detailed analysis of miRNA expression patterns and transcriptional control regions.
Conflict of interest statement
The authors declare no competing interests.
Figures
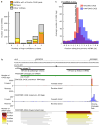
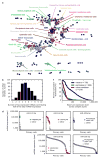
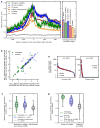
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Li M, Izpisua Belmonte JC. Roles for noncoding RNAs in cell-fate determination and regeneration. Nat Struct Mol Biol. 2015;22:2–4. - PubMed
-
- Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16:279–294. - PubMed
-
- Hasuwa H, Ueda J, Ikawa M, Okabe M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 2013;341:71–73. - PubMed
MeSH terms
Substances
Grants and funding
- BBS/E/D/20211550/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P30 CA010815/CA/NCI NIH HHS/United States
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211552/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I024801/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

