A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization
- PMID: 28829947
- PMCID: PMC5575849
- DOI: 10.1016/j.devcel.2017.07.024
A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization
Abstract
Regulated protein-protein interactions are critical for cell signaling, differentiation, and development. For the study of dynamic regulation of protein interactions in vivo, there is a need for techniques that can yield time-resolved information and probe multiple protein binding partners simultaneously, using small amounts of starting material. Here we describe a single-cell protein interaction assay. Single-cell lysates are generated at defined time points and analyzed using single-molecule pull-down, yielding information about dynamic protein complex regulation in vivo. We established the utility of this approach by studying PAR polarity proteins, which mediate polarization of many animal cell types. We uncovered striking regulation of PAR complex composition and stoichiometry during Caenorhabditis elegans zygote polarization, which takes place in less than 20 min. PAR complex dynamics are linked to the cell cycle by Polo-like kinase 1 and govern the movement of PAR proteins to establish polarity. Our results demonstrate an approach to study dynamic biochemical events in vivo.
Keywords: C. elegans; PAR proteins; PAR-3; PLK-1; SiMPull; cell polarity; cortical flow; sc-SiMPull; single-cell biochemistry.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



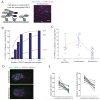
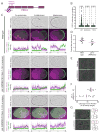
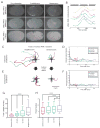

Comment in
-
Protein Clustering Shapes Polarity Protein Gradients.Dev Cell. 2017 Aug 21;42(4):309-311. doi: 10.1016/j.devcel.2017.08.006. Dev Cell. 2017. PMID: 28829939
References
-
- Assémat E, Bazellières E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–630. - PubMed
-
- Beers M, Kemphues K. Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. Development. 2006;133:3745–3754. - PubMed
-
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003a;13:1330–1334. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

