MERS-CoV Accessory ORFs Play Key Role for Infection and Pathogenesis
- PMID: 28830941
- PMCID: PMC5565963
- DOI: 10.1128/mBio.00665-17
MERS-CoV Accessory ORFs Play Key Role for Infection and Pathogenesis
Abstract
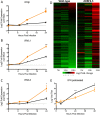
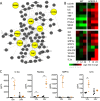
While dispensable for viral replication, coronavirus (CoV) accessory open reading frame (ORF) proteins often play critical roles during infection and pathogenesis. Utilizing a previously generated mutant, we demonstrate that the absence of all four Middle East respiratory syndrome CoV (MERS-CoV) accessory ORFs (deletion of ORF3, -4a, -4b, and -5 [dORF3-5]) has major implications for viral replication and pathogenesis. Importantly, attenuation of the dORF3-5 mutant is primarily driven by dysregulated host responses, including disrupted cell processes, augmented interferon (IFN) pathway activation, and robust inflammation. In vitro replication attenuation also extends to in vivo models, allowing use of dORF3-5 as a live attenuated vaccine platform. Finally, examination of ORF5 implicates a partial role in modulation of NF-κB-mediated inflammation. Together, the results demonstrate the importance of MERS-CoV accessory ORFs for pathogenesis and highlight them as potential targets for surveillance and therapeutic treatments moving forward.IMPORTANCE The initial emergence and periodic outbreaks of MERS-CoV highlight a continuing threat posed by zoonotic pathogens to global public health. In these studies, mutant virus generation demonstrates the necessity of accessory ORFs in regard to MERS-CoV infection and pathogenesis. With this in mind, accessory ORF functions can be targeted for both therapeutic and vaccine treatments in response to MERS-CoV and related group 2C coronaviruses. In addition, disruption of accessory ORFs in parallel may offer a rapid response platform to attenuation of future emergent strains based on both SARS- and MERS-CoV accessory ORF mutants.
Keywords: MERS-CoV; SARS-CoV; coronavirus; live vector vaccines; reverse genetics.
Copyright © 2017 Menachery et al.
Figures

References
-
- WHO 17 September 2015. Middle East respiratory syndrome coronavirus (MERS-CoV)—Saudi Arabia. World Healthy Organization, Geneva, Switzerland: http://www.who.int/csr/don/17-september-2015-mers-saudi-arabia/en/.
-
- Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, Navarrete-Macias I, Liang E, Wells H, Hicks A, Petrosov A, Byarugaba DK, Debbink K, Dinnon KH, Scobey T, Randell SH, Yount BL, Cranfield M, Johnson CK, Baric RS, Lipkin WI, Mazet JAK. 2017. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8:e00373-17. doi:10.1128/mBio.00373-17. - DOI - PMC - PubMed
-
- Haagmans BL, van den Brand JM, Raj VS, Volz A, Wohlsein P, Smits SL, Schipper D, Bestebroer TM, Okba N, Fux R, Bensaid A, Solanes Foz D, Kuiken T, Baumgärtner W, Segalés J, Sutter G, Osterhaus AD. 2016. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science 351:77–81. doi:10.1126/science.aad1283. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
