Fungal lifestyle reflected in serine protease repertoire
- PMID: 28831173
- PMCID: PMC5567314
- DOI: 10.1038/s41598-017-09644-w
Fungal lifestyle reflected in serine protease repertoire
Abstract
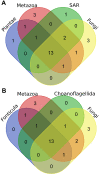
Fungi are able to switch between different lifestyles in order to adapt to environmental changes. Their ecological strategy is connected to their secretome as fungi obtain nutrients by secreting hydrolytic enzymes to their surrounding and acquiring the digested molecules. We focus on fungal serine proteases (SPs), the phylogenetic distribution of which is barely described so far. In order to collect a complete set of fungal proteases, we searched over 600 fungal proteomes. Obtained results suggest that serine proteases are more ubiquitous than expected. From 54 SP families described in MEROPS Peptidase Database, 21 are present in fungi. Interestingly, 14 of them are also present in Metazoa and Viridiplantae - this suggests that, except one (S64), all fungal SP families evolved before plants and fungi diverged. Most representatives of sequenced eukaryotic lineages encode a set of 13-16 SP families. The number of SPs from each family varies among the analysed taxa. The most abundant are S8 proteases. In order to verify hypotheses linking lifestyle and expansions of particular SP, we performed statistical analyses and revealed previously undescribed associations. Here, we present a comprehensive evolutionary history of fungal SP families in the context of fungal ecology and fungal tree of life.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Comment in
-
Commentary: Fungal lifestyle reflected in serine protease repertoire.Front Microbiol. 2018 Mar 13;9:467. doi: 10.3389/fmicb.2018.00467. eCollection 2018. Front Microbiol. 2018. PMID: 29593699 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

