Quantitative profiling of innate immune activation by viral infection in single cells
- PMID: 28831492
- PMCID: PMC5603422
- DOI: 10.1039/c7ib00082k
Quantitative profiling of innate immune activation by viral infection in single cells
Abstract
Cells infected by viruses can exhibit diverse patterns of viral and cellular gene expression. The patterns arise in part from the stochastic or noisy reaction kinetics associated with the small number of genomes, enzymes, and other molecules that typically initiate virus replication and activate cellular anti-viral defenses. It is not known what features, if any, of the early viral or cellular gene expression correlate with later processes of viral replication or cell survival. Here we used two fluorescent reporters to visualize innate immune activation of human prostate cancer (PC3) cells against infection by vesicular stomatitis virus. The cells were engineered to express green-fluorescent protein under control of the promoter for IFIT2, an interferon-sensitive component of the anti-viral response, while red-fluorescent protein was expressed as a byproduct of virus infection. To isolate and quantitatively analyze single-cells, we used a unique microwell array device and open-source image processing software. Kinetic analysis of viral and cellular reporter profiles from hundreds of cells revealed novel relationships between gene expression and the outcome of infection. Specifically, the relative timing rather than the magnitude of the viral gene expression and innate immune activation correlated with the infection outcome. Earlier viral or anti-viral gene expression favored or hindered virus growth, respectively. Further, analysis of kinetic parameters estimated from these data suggests a trade-off between robust antiviral signaling and cell death, as indicated by a higher rate of detectable cell lysis in infected cells with a detectable immune response. In short, cells that activate an immune response lyse at a higher rate. More broadly, we demonstrate how the intrinsic heterogeneity of individual cell behaviors can be exploited to discover features of viral and host gene expression that correlate with single-cell outcomes, which will ultimately impact whether or not infections spread.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures


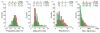

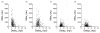

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

