Cancer-associated fecal microbial markers in colorectal cancer detection
- PMID: 28833079
- PMCID: PMC5697688
- DOI: 10.1002/ijc.31011
Cancer-associated fecal microbial markers in colorectal cancer detection
Abstract
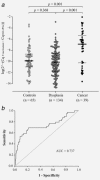
Colorectal cancer (CRC) is the second most common cause of cancer death in the western world. An effective screening program leading to early detection of disease would severely reduce the mortality of CRC. Alterations in the gut microbiota have been linked to CRC, but the potential of microbial markers for use in CRC screening has been largely unstudied. We used a nested case-control study of 238 study subjects to explore the use of microbial markers for clbA+ bacteria harboring the pks pathogenicity island, afa-C+ diffusely adherent Escherichia coli harboring the afa-1 operon, and Fusobacterium nucleatum in stool as potential screening markers for CRC. We found that individual markers for clbA+ bacteria and F. nucleatum were more abundant in stool of patients with CRC, and could predict cancer with a relatively high specificity (81.5% and 76.9%, respectively) and with a sensitivity of 56.4% and 69.2%, respectively. In a combined test of clbA+ bacteria and F. nucleatum, CRC was detected with a specificity of 63.1% and a sensitivity of 84.6%. Our findings support a potential value of microbial factors in stool as putative noninvasive biomarkers for CRC detection. We propose that microbial markers may represent an important future screening strategy for CRC, selecting patients with a "high-risk" microbial pattern to other further diagnostic procedures such as colonoscopy.
Keywords: F. nucleatum; clbA; colorectal cancer; gut microbiota; screening; stool.
© 2017 The Authors International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

