Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals
- PMID: 28834331
- PMCID: PMC5635170
- DOI: 10.1002/mbo3.509
Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals
Abstract
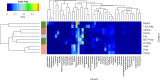
In this pilot study, we determined the core fecal microbiota composition and overall microbiota diversity of domesticated herbivorous animals of three digestion types: hindgut fermenters, ruminants, and monogastrics. The 42 animals representing 10 animal species were housed on a single farm in Ireland and all the large herbivores consumed similar feed, harmonizing two of the environmental factors that influence the microbiota. Similar to other mammals, the fecal microbiota of all these animals was dominated by the Firmicutes and Bacteroidetes phyla. The fecal microbiota spanning all digestion types comprised 42% of the genera identified. Host phylogeny and, to a lesser extent, digestion type determined the microbiota diversity in these domesticated herbivores. This pilot study forms a platform for future studies into the microbiota of nonbovine and nonequine domesticated herbivorous animals.
Keywords: diet; herbivore; hindgut; microbiota; monogastric; ruminant.
© 2017 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures

References
-
- Ahn, J.‐H. , Kim, B.‐Y. , Song, J. , & Weon, H.‐Y. (2012). Effects of PCR cycle number and DNA polymerase type on the 16S rRNA gene pyrosequencing analysis of bacterial communities. Journal of Microbiology, 50, 1071–1074. - PubMed
-
- Barker, C. J. , Gillett, A. , Polkinghorne, A. , & Timms, P. (2013). Investigation of the koala (Phascolarctos cinereus) hindgut microbiome via 16S pyrosequencing. Veterinary Microbiology, 167, 554–564. - PubMed
-
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

