From phenologs to silent suppressors: Identifying potential therapeutic targets for human disease
- PMID: 28834577
- PMCID: PMC5690827
- DOI: 10.1002/mrd.22880
From phenologs to silent suppressors: Identifying potential therapeutic targets for human disease
Abstract
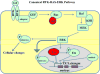
Orthologous phenotypes, or phenologs, are seemingly unrelated phenotypes generated by mutations in a conserved set of genes. Phenologs have been widely observed and accepted by those who study model organisms, and allow one to study a set of genes in a model organism to learn more about the function of those genes in other organisms, including humans. At the cellular and molecular level, these conserved genes likely function in a very similar mode, but are doing so in different tissues or cell types and can result in different phenotypic effects. For example, the RAS-RAF-MEK-MAPK pathway in animals is a highly conserved signaling pathway that animals adopted for numerous biological processes, such as vulval induction in Caenorhabditis elegans and cell proliferation in mammalian cells; but this same gene set has been co-opted to function in a variety of cellular contexts. In this review, I give a few examples of how suppressor screens in model organisms (with a emphasis on C. elegans) can identify new genes that function in a conserved pathway in many other organisms. I also demonstrate how the identification of such genes can lead to important insights into mammalian biology. From such screens, an occasional silent suppressor that does not cause a phenotype on its own is found; such suppressors thus make for good candidates as therapeutic targets.
Keywords: rare disease; resiliency; suppression screen.
© Published 2017. This article is a U.S. Government work and is in the public domain in the USA.
Figures


References
-
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53(4):549–554. - PubMed
-
- Altmann HM, Tester DJ, Will ML, Middha S, Evans JM, Eckloff BW, Ackerman MJ. Homozygous/Compound Heterozygous Triadin Mutations Associated With Autosomal-Recessive Long-QT Syndrome and Pediatric Sudden Cardiac Arrest: Elucidation of the Triadin Knockout Syndrome. Circulation. 2015;131(23):2051–2060. - PubMed
-
- Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401(6751):386–389. - PubMed
-
- Beitel GJ, Clark SG, Horvitz HR. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348(6301):503–509. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

