Genetic architecture of atherosclerosis dissected by QTL analyses in three F2 intercrosses of apolipoprotein E-null mice on C57BL6/J, DBA/2J and 129S6/SvEvTac backgrounds
- PMID: 28837567
- PMCID: PMC5570285
- DOI: 10.1371/journal.pone.0182882
Genetic architecture of atherosclerosis dissected by QTL analyses in three F2 intercrosses of apolipoprotein E-null mice on C57BL6/J, DBA/2J and 129S6/SvEvTac backgrounds
Abstract
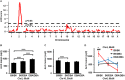
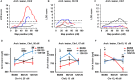
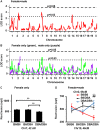
Quantitative trait locus (QTL) analyses of intercross populations between widely used mouse inbred strains provide a powerful approach for uncovering genetic factors that influence susceptibility to atherosclerosis. Epistatic interactions are common in complex phenotypes and depend on genetic backgrounds. To dissect genetic architecture of atherosclerosis, we analyzed F2 progeny from a cross between apolipoprotein E-null mice on DBA/2J (DBA-apoE) and C57BL/6J (B6-apoE) genetic backgrounds and compared the results with those from two previous F2 crosses of apolipoprotein E-null mice on 129S6/SvEvTac (129-apoE) and DBA-apoE backgrounds, and B6-apoE and 129-apoE backgrounds. In these round-robin crosses, in which each parental strain was crossed with two others, large-effect QTLs are expected to be detectable at least in two crosses. On the other hand, observation of QTLs in one cross only may indicate epistasis and/or absence of statistical power. For atherosclerosis at the aortic arch, Aath4 on chromosome (Chr)2:66 cM follows the first pattern, with significant QTL peaks in (DBAx129)F2 and (B6xDBA)F2 mice but not in (B6x129)F2 mice. We conclude that genetic variants unique to DBA/2J at Aath4 confer susceptibility to atherosclerosis at the aortic arch. A similar pattern was observed for Aath5 on chr10:35 cM, verifying that the variants unique to DBA/2J at this locus protect against arch plaque development. However, multiple loci, including Aath1 (Chr1:49 cM), and Aath2 (Chr1:70 cM) follow the second type of pattern, showing significant peaks in only one of the three crosses (B6-apoE x 129-apoE). As for atherosclerosis at aortic root, the majority of QTLs, including Ath29 (Chr9:33 cM), Ath44 (Chr1:68 cM) and Ath45 (Chr2:83 cM), was also inconsistent, being significant in only one of the three crosses. Only the QTL on Chr7:37 cM was consistently suggestive in two of the three crosses. Thus QTL analysis of round-robin crosses revealed the genetic architecture of atherosclerosis.
Conflict of interest statement
Figures

References
-
- Kessler T, Vilne B, Schunkert H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol Med. 2016;8(7): 688–701. doi: 10.15252/emmm.201506174 - DOI - PMC - PubMed
-
- Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77: 1–15. doi: 10.1086/431656 - DOI - PMC - PubMed
-
- Maeda N, Johnson L, Kim S, Hagaman J, Friedman M, Reddick R. Anatomical differences and atherosclerosis in apolipoprotein E-deficient mice with 129/SvEv and C57BL/6 genetic backgrounds. Atherosclerosis. 2007;195: 75–82. doi: 10.1016/j.atherosclerosis.2006.12.006 - DOI - PMC - PubMed
-
- Kayashima Y, Tomita H, Zhilicheva S, Kim S, Kim HS, Bennett BJ, et al. Quantitative trait loci affecting atherosclerosis at the aortic root identified in an intercross between DBA2J and 129S6 apolipoprotein E-null mice. PLoS One. 2014;9: e88274 doi: 10.1371/journal.pone.0088274 - DOI - PMC - PubMed
-
- Tomita H, Zhilicheva S, Kim S, Maeda N. Aortic arch curvature and atherosclerosis have overlapping quantitative trait loci in a cross between 129S6/SvEvTac and C57BL/6J apolipoprotein E-null mice. Circ Res. 2010;106: 1052–1060. doi: 10.1161/CIRCRESAHA.109.207175 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

