Mouse Genome Informatics (MGI) Resource: Genetic, Genomic, and Biological Knowledgebase for the Laboratory Mouse
- PMID: 28838066
- PMCID: PMC5886341
- DOI: 10.1093/ilar/ilx013
Mouse Genome Informatics (MGI) Resource: Genetic, Genomic, and Biological Knowledgebase for the Laboratory Mouse
Abstract
The Mouse Genome Informatics (MGI) Resource supports basic, translational, and computational research by providing high-quality, integrated data on the genetics, genomics, and biology of the laboratory mouse. MGI serves a strategic role for the scientific community in facilitating biomedical, experimental, and computational studies investigating the genetics and processes of diseases and enabling the development and testing of new disease models and therapeutic interventions. This review describes the nexus of the body of growing genetic and biological data and the advances in computer technology in the late 1980s, including the World Wide Web, that together launched the beginnings of MGI. MGI develops and maintains a gold-standard resource that reflects the current state of knowledge, provides semantic and contextual data integration that fosters hypothesis testing, continually develops new and improved tools for searching and analysis, and partners with the scientific community to assure research data needs are met. Here we describe one slice of MGI relating to the development of community-wide large-scale mutagenesis and phenotyping projects and introduce ways to access and use these MGI data. References and links to additional MGI aspects are provided.
Keywords: database; genetics; genomics; human disease model; informatics; model organism; mouse; phenotypes.
© The Author 2017. Published by Oxford University Press.
Figures
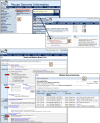
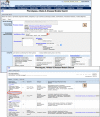
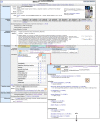
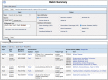
References
-
- Accili D. 2004. A note of caution on the Knockout Mouse Project. Nat Genet 36:1132. - PubMed
-
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. - PMC - PubMed
-
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Juettemann T, Keenan S, Laird MR, Lavidas I, Maurel T, McLaren W, Moore B, Murphy DN, Nag R, Newman V, Nuhn M, Ong CK, Parker A, Patricio M, Riat HS, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Wilder SP, Zadissa A, Kostadima M, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Cunningham F, Yates A, Zerbino DR, Flicek P. 2017. Ensembl 2017. Nucleic Acids Res 45:D635–D642. - PMC - PubMed
-
- Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF. 2010. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol 477:109–123. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

