Redox regulation of plant stem cell fate
- PMID: 28838936
- PMCID: PMC5623875
- DOI: 10.15252/embj.201695955
Redox regulation of plant stem cell fate
Abstract
Despite the importance of stem cells in plant and animal development, the common mechanisms of stem cell maintenance in both systems have remained elusive. Recently, the importance of hydrogen peroxide (H2O2) signaling in priming stem cell differentiation has been extensively studied in animals. Here, we show that different forms of reactive oxygen species (ROS) have antagonistic roles in plant stem cell regulation, which were established by distinct spatiotemporal patterns of ROS-metabolizing enzymes. The superoxide anion (O2·-) is markedly enriched in stem cells to activate WUSCHEL and maintain stemness, whereas H2O2 is more abundant in the differentiating peripheral zone to promote stem cell differentiation. Moreover, H2O2 negatively regulates O2·- biosynthesis in stem cells, and increasing H2O2 levels or scavenging O2·- leads to the termination of stem cells. Our results provide a mechanistic framework for ROS-mediated control of plant stem cell fate and demonstrate that the balance between O2·- and H2O2 is key to stem cell maintenance and differentiation.
Keywords: WUSCHEL; plant stem cell; reactive oxygen species; superoxide anion; superoxide dismutase.
© 2017 The Authors.
Figures
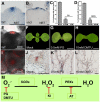
- A, B
NBT staining of the WT and clv3 mutant shows that is highly accumulated in the stem cells. Scale bars, 50 μm. NBT, nitroblue tetrazolium.
- C, D
The percentages of plants with the first pair of true leaves after 7 days after germination (DAG) on media with different PG (C) or DMTU contents (D). More than 200 plants were counted for each treatment. Mean ± SD. ***P < 0.001, Student's t‐test. PG, n‐propyl gallate; DMTU, N,N′‐dimethylthiourea; SD, standard deviation.
- E
The wild‐type inflorescence with dihydroethidium (DHE) staining shows that is highly accumulated in stem cells using longitudinal sections. Scale bar, 50 μm. DHE, dihydroethidium.
- F–H
Seven DAG of wild‐type seedlings on mock medium (F), 0.5 mM PG medium (G), and 10 mM DMTU medium (H). Scale bars, 500 μm.
- I
DHE staining of the wild‐type inflorescence using transverse sections. Scale bar, 50 μm.
- J–L
CLV3 expression patterns of the 7 DAG wild type on mock medium (J), 0.5 mM PG medium (K), and 10 mM DMTU medium (L). All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- M
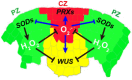
Diagram of ROS metabolism in plants. KI, potassium iodide; AT, amino‐1,2,4‐triazole.
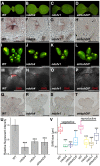
- A–D
The top view of 7‐day‐old seedlings of the wild‐type plant (A) and the ndufs4 (B), ndufv1 (C), and atrbohD/F (D) mutants. Scale bars, 500 μm.
- E–H
CLV3 expression patterns of the 7‐day‐old wild‐type plant (E) and the ndufs4 (F), ndufv1 (G), and atrbohD/F (H) mutants. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- I–L
Top view of inflorescence in the wild‐type plant (I) and in the ndufs4 (J), ndufv1 (K), and atrbohD/F (L) superoxide‐deficient mutants shows that there are fewer floral buds than the wild‐type plant. Scale bars, 1 mm.
- M–P
DHE staining of the wild‐type (M), ndufs4 (N), ndufv1 (O), and atrbohD/F (P) inflorescences. Scale bars, 50 μm.
- Q–T
CLV3 expression patterns in the wild‐type plant (Q) and the ndufs4 (R), ndufv1(S), and atrbohD/F (T) mutants show reduced SAM sizes and CLV3 expression domains in the mutants at the reproductive stage. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- U
Quantification of DHE fluorescent intensity in (M–P) (WT, n = 10; ndufs4, n = 7; ndufv1 and atrbohD/F, n = 8; ± SD). ***P < 0.001, Student's t‐test.
- V
Quantification of the SAM size in the wild‐type plant and the ndufs4, ndufv1, and atrbohD/F mutants in both the vegetative and reproductive stages. In the vegetative stage: WT and ndufv1, n = 12, ndufs4 and atrbohD/F, n = 11, in the reproductive stage: WT, n = 25, ndufv1, ndufs4, and atrbohD/F, n = 16. Upper and lower limits of box correspond to upper Q3 and lower Q1 quartiles, the horizontal line within the box represent the data medians, the single value (open square) plotted inside the box indicate the data means, error bars indicating minimum to maximum range, whiskers indicate data range within 1.5× of the interquartile range and outliers are not shown. ***P < 0.001, Student's t‐test.

- A–D
WUS expression patterns in 7‐day‐old seedlings of the wild‐type plant (A) and the ndufs4 (B), ndufv1 (C), and atrbohD/F (D) mutants. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- E–H
WUS expression patterns of the wild‐type plant (E) and the ndufs4 (F), ndufv1 (G), and atrbohD/F (H) mutants at the reproductive stage. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- I
Quantitative measurements of the WUS and CLV3 expression levels in the wild‐type plant and the ndufs4, ndufv1, and atrbohD/F mutants in the vegetative stages. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001, Student's t‐test.
- J
Quantitative measurements of the WUS and CLV3 expression levels in the wild‐type plant and the ndufs4, ndufv1, and atrbohD/F mutants in the reproductive stages. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.

- A, B
DHE staining in the CLV3::ALR, ALA::amiRNA‐NDUFS4 transgenic plants with (B) and without (A) ethanol induction. Scale bars, 50 μm.
- C
Quantification of DHE fluorescent intensity in CLV3::ALR, ALA::amiRNA‐NDUFS4 transgenic plants with or without ethanol induction (n = 7, ± SD). **P < 0.01, Student's t‐test.
- D–F
Quantitative measurement of NDUFS4 (D), WUS (E), and CLV3 (F) expression levels in CLV3::ALR, ALA::amiRNA‐NDUFS4 and CLV3::ALR, ALA::GUS plants with or without ethanol induction. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001, Student's t‐test.

- A–F
The spatiotemporal expression patterns of six SODs in the wild‐type plants, including FSD2 (A), CSD1 (B), CSD3 (C), MSD1 (D), CSD2 (E), and FSD1 (F). Scale bars, 50 μm.
- G–I
The phenotypes of the wild‐type plant (G), CLV3::FSD2 (H, 6 out of 39 transgenic T1 plants showed the meristem termination phenotypes) and UBQ10::FSD2 (I, 7 out of 36 transgenic T1 plants showed the meristem termination phenotypes). Scale bars, 1 mm.
- J–L
WUS expression patterns in the wild‐type plant (J), CLV3::FSD2 (K), and UBQ10::FSD2 (L) transgenic plants. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- M–O
CLV3 expression patterns in the wild‐type (M), CLV3::FSD2 (N), and UBQ10::FSD2 (O) transgenic plants. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.

- A, B
CSD1 expression patterns in the SAM at the reproductive stage (A1–A4, serial longitudinal sections; B, transverse section). Scale bars, 50 μm.
- C, D
CSD2 expression patterns in the SAM at the reproductive stage (C1–C4, serial longitudinal sections; D, transverse section). Scale bars, 50 μm.
- E, F
CSD3 expression patterns in the SAM at the reproductive stage (E1–E4, serial longitudinal sections; F, transverse section). Scale bars, 50 μm.
- G, H
FSD2 expression patterns in the SAM at the reproductive stage (G1–G4, serial longitudinal sections; H, transverse section). Scale bars, 50 μm.
- I, J
MSD1 expression patterns in the SAM at the reproductive stage (I1–I4, serial longitudinal sections; J, transverse section). Scale bars, 50 μm.
- K, L
FSD1 expression patterns in the SAM at the reproductive stage (K1–K4, serial longitudinal sections; L, transverse section). Scale bars, 50 μm.
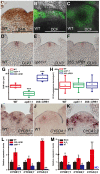
- A–C
Wild‐type inflorescences stained with DAB (A) and DCF (B, longitudinal section; C, transverse section) show that H2O2 is more abundant in the PZ. Scale bars, 50 μm. DAB, 3,3′‐diaminobenzidine; DCF, 2′,7′‐dichlorofluorescin diacetate.
- D–F
CLV3 expression patterns of the wild‐type (D), upb1‐1‐mutant (E), and 35S:UPB1 transgenic plants (F). All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
- G
Quantification of the SAM size in the wild‐type, upb1‐1 mutant, and 35S:UPB1 transgenic plants. Upper and lower limits of box correspond to upper Q3 and lower Q1 quartiles, the horizontal line within the box represent the data medians, the single value (square) plotted inside the box indicate the data means, error bars indicating minimum to maximum range, whiskers indicate data range within 1.5× of the interquartile range and outliers are not shown. For each genotype, n = 12, ***P < 0.001, Student's t‐test.
- H
Quantification of the CLV3 expression domains in the wild‐type, upb1‐1, and 35S::UPB1 plants. Boxplot representation is as described in panel (G). For each genotype, n = 12.
- I–K
The expression patterns of CYCB1;1 (I), CYCD4;1 (J), and CYCA3;3 (K) in the wild‐type SAM. Scale bars, 50 μm.
- L
Quantitative measurement of the CYCB1;1, CYCD4;1, and CYCA3;3 expression levels in the wild‐type, upb1‐1, and 35S::UPB1 plants. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.
- M
Quantitative measurement of the CYCB1;1, CYCD4;1, and CYCA3;3 expression levels under the 16‐h H2O2 or KI treatments. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test. ND, not detected.
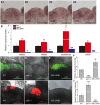
- A
UPB1 expression patterns in the wild‐type SAM (A1–A4, serial longitudinal sections). Scale bars, 50 μm.
- B
Three stem cell‐specific peroxidases, including PRX33, PRX52, and AT5G64100, were repressed by UPB1 in the SAM. The Per57 was used as a positive control. Mean ± SD with three independent biological replicates. *P < 0.05 and **P < 0.01, Student's t‐test. ND, not detected.
- C–E
DCF staining in the wild‐type (C), upb1‐1 (D), and 35S::UPB1 (E) plants. Scale bars, 50 μm.
- F
Quantification of the fluorescent intensity of DCF in (C–E). N = 11, ± SD; **P < 0.01 and ***P < 0.001, Student's t‐test.
- G–I
DHE staining in the wild‐type (G), upb1‐1 (H), and 35S::UPB1 (I) plants. Scale bars, 50 μm.
- J
Quantification of the fluorescent intensity of DHE in (G–I). WT and upb1‐1, n = 15, ± SD; 35S::UPB1, n = 12, ± SD; ***P < 0.001, Student's t‐test.
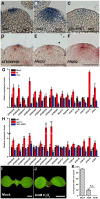
- A–C
distribution patterns in the SAMs of the wild‐type (A), upb1‐1‐mutant (B), and 35S:UPB1 transgenic plants (C). Scale bars, 50 μm.
- D–F
Peroxidase gene expression patterns in the wild‐type SAM, including AT5g64100 (D), PRX33 (E), and PRX52 (F). Scale bars, 50 μm.
- G
Quantitative measurement of the NADH dehydrogenase, NADPH oxidase, and alternative oxidase expression levels in the SAM under the 16‐h H2O2 or KI treatments. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.
- H
Quantitative measurement of the NADH dehydrogenase, NADPH oxidase, and alternative oxidase expression levels in the SAMs of the wild‐type, upb1‐1, and 35S::UPB1 plants. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.
- I, J
Seven days after the germination of the wild‐type seedlings in the mock (I) and H2O2‐containing media (J). Scale bars, 500 μm.
- K
The percentage of wild‐type plants with the first pair of true leaves after 7 days of germination on media with different H2O2 concentrations. More than 200 plants were counted for each treatment. Mean ± SD. ***P < 0.001, Student's t‐test.
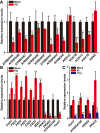
Quantitative measurement of NADPH oxidase and alternative oxidase expression levels by qRT–PCR in response to the short‐term (16‐h) treatment with PG. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.
The expression levels of the SODs and peroxidases in response to the short‐term (16‐h) treatment with PG. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t‐test.
The expression levels of the peroxidases in response to the short‐term (16‐h) treatment with KI or H2O2. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.

- A
WUS expression levels in 11‐day‐old wild‐type seedlings under the short‐term (16‐h) treatments with DMTU, PG, MV, H2O2, AT, and KI were measured by qRT–PCR. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, Student's t‐test.
- B–E
DHE staining (top row) and the WUS protein (bottom row) in the 11‐day‐old wild‐type seedlings under the short‐term (16‐h) treatments with mock (B), DMTU (C), PG (D), and MV (E). The dotted yellow line represents the L1 layer of the meristem. Scale bars, 50 μm.
- F–I
DCF staining (top row) and the WUS protein (bottom row) in 11‐day‐old wild‐type seedlings under the short‐term (16‐h) treatments with mock (F), H2O2 (G), AT (H), and KI (I). The dotted yellow line represents the L1 layer of the meristem. Scale bars, 50 μm.
- J
Quantification of the fluorescent intensity of DHE in (B–E) (mock and PG, n = 10, ± SD; DMTU and MV, n = 7, ± SD). *P < 0.05 and ***P < 0.001, Student's t‐test.
- K
Quantification of the fluorescent intensity of DCF in (F–I) (mock and KI, n = 10, ± SD; AT, n = 6, ± SD; H2O2, n = 12, ± SD). **P < 0.01 and ***P < 0.001, Student's t‐test.
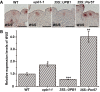
WUS expression patterns in the wild‐type, upb1‐1, 35::UPB1, and 35S::Per57 plants. All in situ hybridizations were performed in the same time and same conditions. Scale bars, 50 μm.
WUS expression levels in the wild‐type, upb1‐1, 35::UPB1, and 35S::Per57 plants measured by qRT–PCR. Mean ± SD with three independent biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t‐test.
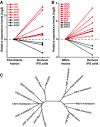
The expression levels of SOD1, SOD2, and SOD3 in human fibroblasts were downregulated during the process of iPSCs induction by treatment with c‐Myc and Oct4, whereas six peroxidases were upregulated. The transcriptome data were obtained from the previous study (Soldner et al, 2009).
SOD1, SOD2, and SOD3 in mouse were downregulated during the process of iPSCs induction of neural stem cells (NSCs) with the Oct4 treatment, whereas nine peroxidases were upregulated in the same scenario. The transcriptome data were obtained from the previous study (Kim et al, 2009).
Phylogenetic analysis of SODs among Arabidopsis, Mus, Homo, and Drosophila. SOD1 and SOD3 in animal were highly conserved with CSDs in Arabidopsis, whereas FSDs and MSD1 in Arabidopsis were more close to SOD2 in animals.
References
-
- Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 - PubMed
-
- Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53: 1331–1341 - PubMed
-
- Andreyev AI, Kushnareva YE, Starkov AA (2005) Mitochondrial metabolism of reactive oxygen species. Biochem (Mosc) 70: 200–214 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

