Activation-tagging in indica rice identifies a novel transcription factor subunit, NF-YC13 associated with salt tolerance
- PMID: 28839256
- PMCID: PMC5570948
- DOI: 10.1038/s41598-017-10022-9
Activation-tagging in indica rice identifies a novel transcription factor subunit, NF-YC13 associated with salt tolerance
Abstract
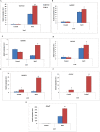
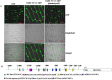
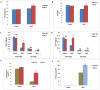
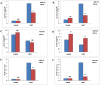
Nuclear factor Y (NF-Y) is a heterotrimeric transcription factor with three distinct NF-YA, NF-YB and NF-YC subunits. It plays important roles in plant growth, development and stress responses. We have reported earlier on development of gain-of-function mutants in an indica rice cultivar, BPT-5204. Now, we screened 927 seeds from 70 Ac/Ds plants for salinity tolerance and identified one activation-tagged salt tolerant DS plant (DS-16, T3 generation) that showed enhanced expression of a novel 'histone-like transcription factor' belonging to rice NF-Y subfamily C and was named as OsNF-YC13. Localization studies using GFP-fusion showed that the protein is localized to nucleus and cytoplasm. Real time expression analysis confirmed upregulation of transcript levels of OsNF-YC13 during salt treatment in a tissue specific manner. Biochemical and physiological characterization of the DS-16 revealed enhanced K+/Na+ ratio, proline content, chlorophyll content, enzymes with antioxidant activity etc. DS-16 also showed transcriptional up-regulation of genes that are involved in salinity tolerance. In-silico analysis of OsNF-YC13 promoter region evidenced the presence of various key stress-responsive cis-regulatory elements. OsNF-YC13 subunit alone does not appear to have the capacity for direct transcription activation, but appears to interact with the B- subunits in the process of transactivation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Murphy KST, Durako MJ. Physiological effects of short-term salinity changes on Ruppia maritima. Aquat Bot. 2003;75:293–309. doi: 10.1016/S0304-3770(02)00206-1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

